
Topics and trends in gastroesophageal reflux disease research over the past 60 years: a text mining and network analysis
Highlight box
Key findings
• Current research on gastroesophageal reflux disease (GERD) tends to follow traditional ideas, which limits the scope of our understanding of the disease.
• Part of the factors and relationships that may be significant in GERD have not been thoroughly studied or experimentally validated yet.
What is known and what is new?
• GERD is a common disease with suboptimal clinical outcomes due to the significant heterogeneity in the pathogenesis.
• A comprehensive knowledge network of GERD is created through text mining for the first time, revealing fragmented research and persistent knowledge gaps.
What is the implication, and what should change now?
• Our findings underscore that the conceptualization of GERD interactions is largely anchored in clinical studies, reflecting an inadequate understanding of the underlying mechanisms of the disease and alerting researchers not to stand still.
Introduction
Gastroesophageal reflux disease (GERD) is a major public health burden with an estimated worldwide prevalence of 8–33% (1,2), which not only exerts a considerable economic burden but also diminishes the health-related quality of life for affected individuals (3-5). This complex disease is characterized by symptoms such as heartburn and regurgitation caused by the reflux of gastroduodenal contents entering the esophagus (6).
Acid-suppressing drugs serve as the mainstay treatment for GERD, while up to 50% of cases display a lack of symptom improvement despite the use of proton pump inhibitors (2), with some individuals developing new reflux symptoms and esophageal mucosa damage. The suboptimal clinical outcomes are predominantly attributed to the significant heterogeneity in the pathogenesis (7-9), which remains incompletely understood. Current perspectives posit a multifactorial genesis, implicating dysfunction across various domains, including genetic susceptibility, esophageal motility disorders, injury induced by immune inflammation, as well as peripheral and central sensitization (10-12).
GERD-related research is rapidly expanding (13), but most of it is biased towards one perspective. Scholars tend to focus solely on aspects within their specialized fields, disregarding the interconnection with other domains. As a result, research on the disease remains fragmented, and knowledge gaps persist. To address these issues, it is imperative to conduct a bibliometric analysis of academic literature to map the knowledge domain (13).
In this study, we conducted a comprehensive analysis of the representation and evolution of components in GERD research over the past decades by text mining of abstracts and network analysis. This study aimed to analyze the developmental trends in GERD research field and identify key challenges in constructing the knowledge network.
Methods
The combined approach of text mining and network analysis was applied in this study. Text mining of abstracts was a newly-developed method for bibliometric analysis, which supports knowledge synthesis by identifying important associations between components in complex diseases (14,15). The findings of bibliometric analysis can be concluded as network plots and analyzed mathematically. From the network analysis, we can extract key information and research highlights from a growing set of data. Network theory provides a clear structure for extracting useful insights from growing data sets (16,17). These approaches facilitate a better understanding of the knowledge construction in a field, and better insights of interconnections between the components of complex diseases (18), might help to provide further insights into the field of investigation.
Building a GERD literature abstract library
A systematic literature search on GERD was conducted in PubMed. The search strategy was provided in Appendix 1. Zotero (version 6.0.3, Corporation for Digital Scholarship, USA) was used to automatically download abstracts and metadata (e.g., title, author, publication year, journal, keywords, DOI, and URL), remove duplication and screen literature. Incorrect literature types (i.e., books and documents, Video-audio media, Webcast), articles that not containing human research and unrelated to GERD were excluded. Two trained researchers independently screened data and confirmed findings with a senior professor if there was any inconsistency. The flow chart of screening was provided in Figure S1. The final library was provided in the supplementary table (https://cdn.amegroups.cn/static/public/tgh-24-84-1.csv). Word cloud was generated by RStudio software (version 4.2.2) and package wordcloud2 (19). Output data and stop words were available at https://cdn.amegroups.cn/static/public/tgh-24-84-3.csv.
Building a matrix of components of interest at different functional levels for GERD
Combining domain knowledge and database materials, we used a semi-automatic method to capture components of interest and their synonyms, all of which were manually reviewed by the authors. Considering the characteristics of GERD (14,17,20-26), to create the universe linking genetic, biochemical, cellular, physiological and clinical data, the components of interest were classified into three functional levels: (I) genes and molecules (n=180), containing terms that maintain the integrity of epithelium, regulate inflammation and malignancy, act in the nervous system, constitute digestive juice and gastrointestinal peptide hormone, (II) terms of cytology, histology, and anatomy (n=54), containing cells and tissues that built up the wall of esophagus, join in acid secretion, immunology, carcinogenesis and neural regulation, as well as structures or organs that related to the disease, (III) terms of clinical definition (n=243), comprised epidemiological factors, disease definitions, various symptoms, complications and comorbidities, together with diagnostic techniques and treatments. Synonyms were built based on databases (GeneCards, Malacards, DISEASES, HGNC, Mesh database, Wikipedia, Drug.com) and domain knowledge. A total of 295 synonyms were included, while non-specific synonyms were excluded to minimize false positives. The matrix is provided in the supplementary table (https://cdn.amegroups.cn/static/public/tgh-24-84-2.xlsx).
Scanning components of interest in literature abstracts
MATLAB R2022b was used for automatically scanning the text of abstracts to calculate the information of components of interest and their synonyms. The code was created in a previous study (14). Special characters (such as @, #, ^, etc.) in the abstract library were detected and manually organized as needed when found.
Network analysis visualizing the comparison of nodes and changes over time
Network analysis was used to describe the representation of these components of interest and their changes over time, and to compare the information about these components contained in each stage. In the network, each node represented a component of interest, and edges the co-occurrence of a pair of components. The size and color of a node is determined by the number of edges attached to it (i.e., the components associated with it). The width of the edges is determined by the weights (i.e., the frequency of co-occurrence of a given pair of components), with thicker edges being higher-weight edges.
Results
Overview of the literature abstract database
A total of 18,483 articles were retrieved, and 18,459 articles were obtained after exclusion. Figure 1 shows the overview of the data. The annual publication rate grew exponentially before 2003 and slowed down after 2003. The published journals were found to be 1,854 in number, with a lot of concentration in the field of digestive specialties, while comprehensive journals began to appear in the 9th place. Half of the literature was concentrated in the top 40 journals, and more than 40% of the journals only published one related article. The analysis of all the words from the literature abstract indicated that symptoms and management-related terms are prevalent. The research scope of co-existing diseases is wide, and those involving microscopic biological mechanisms are less significant.
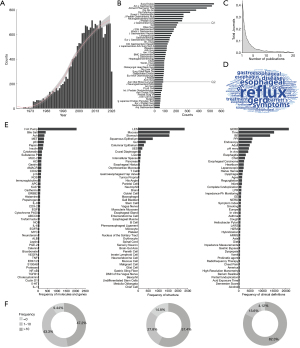
Frequencies of components of interest in the abstracts
We focused on frequencies of components in three different functional levels next. At the molecule level, the proton pump was getting the most attention, emphasizing the acid-related definition of GERD. Constituents of digestive juice, tumor suppressor genes and proto-oncogenes that may associated with Barrett’s esophagus and carcinogenesis, and molecules that act as neurotransmitters were also mentioned, which indicated the diversity of molecule basis of the disease. Among the 54 types of structure, the most prevailing words were the lower esophageal sphincter, mucosa, stomach, and epithelium. Cells like T lymphocytes and the components related to the nervous system were retrieved at a lower frequency, warning that the immunological and neural mechanisms need to be explored badly. The frequency of components related to diagnosis and management was significantly higher than that related to molecules and structures, in terms of which, the most frequently referred to the disease, time-related variables, population, or medication and diagnostic tools. Overall, the frequency of several classic components of interest was significantly higher than that of others, and most of the components appeared in very few papers. Quantitatively, 47.2% of the molecules were mentioned in more than 10 pieces of literature, and for structures and clinical definitions, the number is 57.4% and 82.3% respectively.
Representation of components and their relationship over time
On a temporal scale, the focus of research on molecules has altered greatly over time (Figure 2). In the early stages, gastrointestinal hormones and digestive enzymes were the primary focus. Proton pumps joined the field of study in the 1990s and went on to play a significant role. Following this, a wide range of molecules were studied in detail. These included oncogenes, neurotransmitters and receptors, inflammatory factors such as interleukins, chemokines, and molecules linked to the development of Barrett’s esophagus and cancer. At this point, each category’s high-frequency molecules defined a basic sequence that became completely apparent after the second decade. Though there was a substantial change in the inclusion of new components, and the connection of components in the subsequent decade improved compared to the previous decade, the basic framework remained.
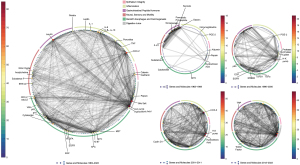
As for structure, the study framework has been maintained from the beginning to the present, with the three vertices of the triangle representing the gut and stomach, the lower esophageal sphincter, and the mucosa and epithelium. The other structures, such as neurons, the brain, synapses, immune cells, malignant cells, and so on, have always received less attention. Gut-brain interaction is a highly-value topic while without enough pieces of literature to support and zoom. Immune cells and malignant cells mildly engage in the circle from 1990–2000.
The definition of diagnosis and management has significantly evolved in both components and connectivity compared to the microscopic definition (Figure 3). However, throughout history, certain key factors have consistently remained dominant. Five key nodes were identified, with several thick edges connecting them. These nodes encompassed (I) disease-related terms such as GERD, erosive esophagitis, non-erosive reflux disease, and Barrett’s esophagus, (II) descriptors of pathological states such as esophagitis, (III) GERD medications including proton pump inhibitors (PPIs), H2 receptor antagonists, and prokinetics, (IV) procedures such as endoscopy, pH meter, and laparoscopy, and (V) time-related variables and different populations. The extra-esophageal symptoms, such as annoying cough and asthma, weren’t recognized until the turn of the century. Considerable words that point out the symptoms, complications, and comorbidities were given in 2001–2011. Early anti-reflux surgeries were performed through laparoscopic intervention in the 1990s and 2000s, but after 2001, less invasive procedures such as radiofrequency therapy and anti-reflux mucosectomy gained popularity. With the advancements in impedance-pH monitoring and manometry, parameters such as acid clearance time and distal latency have also become more widely used.
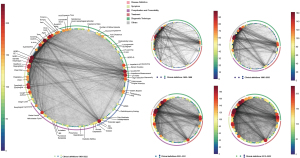
Changes in the network
Table 1 shows the rapid network changes quantitatively. Between 1990 and 2000, the number of unique molecules in the network doubled to 69 and then doubled again in the following decade. The increasing rate slowed down thereafter. During the same period, there was a more notable increase in connectivity, with the number of edges and connectivity doubling from the first decade to the second. However, there was little change in nodes, edges, and connectivity from the second to the third decade. Despite this growth, many molecules’ relevant information was not present in the network. At the structural level, recent years have seen minimal changes. The nodes associated with diagnosis and treatment had the highest cardinality and connectivity, and their development is steady. Compared to the past, there was a commonality in each functional category, and the rapid development time of network attributes is concentrated in the 1990–2000 and 2001–2011 decades. The development in the 2012–2022 decade has slowed down significantly, even stagnated.
Table 1
| Time period | Number nodes | Number edges | Average weight edges | Edge weight entropy | Words entropy | Mean connectivity |
|---|---|---|---|---|---|---|
| Complete set | ||||||
| 1963–1989 | 175 | 2,619 | 678.66 | 11.42 | 0.68 | 14.97 |
| 1990–2000 | 287 | 5,714 | 861.43 | 13.25 | 0.67 | 19.91 |
| 2001–2011 | 383 | 11,341 | 1,187.30 | 17.02 | 0.68 | 29.61 |
| 2012–2022 | 415 | 15,034 | 1,140.94 | 21.21 | 0.68 | 36.23 |
| 1963–2022 | 443 | 19,588 | 1,941.85 | 19.03 | 0.67 | 44.22 |
| Clinical definitions | ||||||
| 1963–1989 | 112 | 1,497 | 893.05 | 8.88 | 0.68 | 13.37 |
| 1990–2000 | 170 | 3,120 | 1,191.57 | 10.21 | 0.67 | 18.35 |
| 2001–2011 | 206 | 5,876 | 1,724.64 | 12.99 | 0.70 | 28.52 |
| 2012–2022 | 231 | 8,359 | 1,586.66 | 16.83 | 0.70 | 36.19 |
| 1963–2022 | 233 | 10,024 | 2,897.02 | 14.63 | 0.68 | 43.02 |
| Definitions of cytology, histology, and anatomy | ||||||
| 1963–1989 | 25 | 69 | 328.99 | 1.33 | 0.60 | 2.76 |
| 1990–2000 | 29 | 86 | 556.98 | 1.79 | 0.62 | 2.97 |
| 2001–2011 | 37 | 141 | 652.48 | 2.28 | 0.63 | 3.81 |
| 2012–2022 | 38 | 164 | 486.59 | 2.75 | 0.63 | 4.32 |
| 1963–2022 | 42 | 228 | 1,069.30 | 2.69 | 0.60 | 5.43 |
| Genes and molecules | ||||||
| 1963–1989 | 31 | 89 | 139.33 | 1.73 | 0.84 | 2.87 |
| 1990–2000 | 69 | 215 | 180.47 | 3.30 | 0.77 | 3.12 |
| 2001–2011 | 122 | 791 | 212.14 | 8.32 | 0.73 | 6.48 |
| 2012–2022 | 134 | 810 | 203.83 | 8.36 | 0.73 | 6.04 |
| 1963–2022 | 152 | 1,407 | 275.69 | 12.37 | 0.74 | 9.26 |
The whole set network was integrated by the three networks from each functional level containing all 443 nodes. Between 1963 and 1989 and the first two decades, a significant increase in component diversity was observed, with the number of nodes increasing by about 64% and 33% in the first and second decades. In the third decade, the number of nodes increased only by 8%. The increase in the number of edges was also slowing, presenting weakness and fatigue over the past decade.
The connectivity and node entropy of the complete set was similar to the network of clinical definitions, implying that the nodes and edges related to clinical definition decide the overall network’s attributes, while those related to molecules and structures contribute less. That means the disease research is dominated by clinical definitions, without enough attraction on underlying factors.
Discussion
To the best of our knowledge, this was the first network analysis of text mining-based study of GERD, presenting the entirety of components and relationships in the GERD research literature. Our findings underscore that the conceptualization of the GERD interactions is largely anchored in clinical field studies, reflects an inadequate understanding of the underlying mechanisms of the disease and alerting researchers not to stand still.
Only a little bibliometric research has explored the status of research on GERD. Zhang et al. collected literatures from 2012 to 2022 found keyword clusters focused on clinical features (13). GERD and disorders of gut-brain interactions have substantial symptom overlaps (27). The bibliometric studies of functional dyspepsia have concluded that overlapping was extensively explored, but again found that basic research has lagged the clinical evidence (28). These studies only used auto-processing software to analyze keywords and headings, without analyzing the literature abstracts or presenting quantitative data, which lost plenty of information. Here, by constructing components of interest at different functional levels and scanning the terms in the literature abstracts, a comprehensive research picture of GERD can be derived. Meanwhile, we use network analysis to show the ebbs and flows of the research hotspots with their connections in the long history and identified general patterns from a huge literature abstract database, finding an urgent need for basic study and brain-gut interactions for the first time.
Diagnosis and management networks predominantly dominate the overall GERD network, signifying that the concentration of GERD research within clinical realms offers valuable insights and treatment avenues for patient management. However, this trend may imply a limited understanding of the disease’s underlying mechanisms. A deeper comprehension of the disease’s nature, the development of more efficacious therapeutic strategies, and addressing the challenges posed by its dynamic changes could substantially enhance patient care and the delivery of more effective treatment modalities.
As expected, our analysis uncovered a consistent expansion in the volume of published literature concerning human GERD over recent decades. This proliferation encompasses both the quantity of GERD-related publications and the comprehensive augmentation of their associated components and relationships. However, a consistent pattern observed in the network nodes and edges within GERD research suggests a prevailing set of assumptions shaping the trajectory of research for at least the past two decades. Researchers today have unprecedented access to a wealth of information, largely owing to the exponential growth of open data facilitated by the Internet. This widespread availability is evident in the consistent annual rise in article indexing within databases such as PubMed/MEDLINE, mirrored in GERD research. Quite many scholars, however, have pointed out the negative impacts of the exploding increase of information in the age of Internet, including information overload and information cocoon (29-32). Yuan et al. found that information homogeneity and value identification are essential elements that influence the formation of researchers’ information cocoon (32). It is noteworthy that this will not be conducive to interdisciplinary academic exchanges or progress of GERD research fields. Current studies on human GERD exhibit tendencies that echo earlier conceptualizations, fostering a conservative structure within our identified network. Many other components and relationships relevant to GERD lack robust experimental validation in basic studies or have not undergone thorough investigation. Our analysis indicates the need for comprehensive testing of these components and relationships.
The historical evolution of GERD research interests is evident. In the early 1990s, emphasis centered on the impact of refluxate, especially gastric acid on the esophageal mucosa. Next, subsequent technological advancements, shifted the spotlight toward the multiple defense and damage mechanisms at the esophageal-gastric junction (10,33-37), leading GERD to a generally acknowledged multifactorial disease. Specifically, within molecular networks, there is a consistent augmentation in both the number of components and the complexity of their interrelationships, notably escalating during the pre-phase. This expansion owes much to the groundwork laid by the biotechnology industry, notably the landmark achievements of the Human Genome Project in the late 1990s and the subsequent Human Proteome Project. Recent years, the extra-esophageal presentations and alternative therapies have gained more have gained more attention (36,38-41). This change in historical context likely influenced research focus and methodological selections. For further research on the GERD components, different research histories and possible scientific biases should be considered to utilize information and technology more effectively.
As previously discussed, the historical evolution and epistemology of GERD research indicate potential revelations of novel components and relationships in future studies. However, concurrently evaluating involved components across various functional levels remains a formidable challenge. Hence, future perspectives in GERD components research should encompass collaborative studies, patient phenotyping, integrated bioinformatics, and expertise, coupled with the long-term, prospective collection of samples from diverse groups involved in its pathogenesis.
The current network analysis gained comprehensive study status of GERD patients which provided novel insights for developing research strategies. However, the following problems still exist in the study. Firstly, our building of an interested matrix may incur a secondary bias towards components of high abundance in the literature. However, it is widely recognized that because the technique used determines the type of relationships found, particularly at the molecular level, the procedures are always prone to bias (42,43). Thus, future knowledge synthesis could implement other approaches to filter components and illustrate different aspects of them than those examined here. Secondly, the source of GERD literature abstract library is merely PubMed. Besides, provisions with conflicts of interest, such as the source of funding, were not considered. Future analyses can distinguish this issue and make comparisons to explain the impact of funding agencies.
Conclusions
The current study utilizes abstract text-mining and network analysis methods to analyze research on GERD for the first time. Our analysis of the massive amounts of information highlights the main challenge to understand this complex disease may be to recognize that knowledge construction in the field remain confined to itself and float at the surface.
Acknowledgments
None.
Footnote
Peer Review File: Available at https://tgh.amegroups.com/article/view/10.21037/tgh-24-84/prf
Funding: The study was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tgh.amegroups.com/article/view/10.21037/tgh-24-84/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- El-Serag HB, Sweet S, Winchester CC, et al. Update on the epidemiology of gastro-oesophageal reflux disease: a systematic review. Gut 2014;63:871-80. [Crossref] [PubMed]
- Delshad SD, Almario CV, Chey WD, et al. Prevalence of Gastroesophageal Reflux Disease and Proton Pump Inhibitor-Refractory Symptoms. Gastroenterology 2020;158:1250-1261.e2. [Crossref] [PubMed]
- The global, regional, and national burden of gastro-oesophageal reflux disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol 2020;5:561-81. [Crossref] [PubMed]
- Sweis R, Fox M. The global burden of gastro-oesophageal reflux disease: more than just heartburn and regurgitation. Lancet Gastroenterol Hepatol 2020;5:519-21. [Crossref] [PubMed]
- Zhang D, Liu S, Li Z, et al. Global, regional and national burden of gastroesophageal reflux disease, 1990-2019: update from the GBD 2019 study. Ann Med 2022;54:1372-84. [Crossref] [PubMed]
- Gyawali CP, Kahrilas PJ, Savarino E, et al. Modern diagnosis of GERD: the Lyon Consensus. Gut 2018;67:1351-62. [Crossref] [PubMed]
- Fass R, Boeckxstaens GE, El-Serag H, et al. Gastro-oesophageal reflux disease. Nat Rev Dis Primers 2021;7:55. [Crossref] [PubMed]
- Katzka DA, Kahrilas PJ. Advances in the diagnosis and management of gastroesophageal reflux disease. BMJ 2020;371:m3786. [Crossref] [PubMed]
- Patti MG. An Evidence-Based Approach to the Treatment of Gastroesophageal Reflux Disease. JAMA Surg 2016;151:73-8. [Crossref] [PubMed]
- Bonfiglio F, Hysi PG, Ek W, et al. A meta-analysis of reflux genome-wide association studies in 6750 Northern Europeans from the general population. Neurogastroenterol Motil 2017; [Crossref] [PubMed]
- Cameron AJ, Lagergren J, Henriksson C, et al. Gastroesophageal reflux disease in monozygotic and dizygotic twins. Gastroenterology 2002;122:55-9. [Crossref] [PubMed]
- Mohammed I, Cherkas LF, Riley SA, et al. Genetic influences in gastro-oesophageal reflux disease: a twin study. Gut 2003;52:1085-9. [Crossref] [PubMed]
- Zhang T, Zhang B, Tian W, et al. Trends in gastroesophageal reflux disease research: A bibliometric and visualized study. Front Med (Lausanne) 2022;9:994534. [Crossref] [PubMed]
- Fernandez ME, Nazar FN, Moine LB, et al. Network Analysis of Inflammatory Bowel Disease Research: Towards the Interactome. J Crohns Colitis 2022;16:1651-62. [Crossref] [PubMed]
- Sintchenko V, Anthony S, Phan XH, et al. A PubMed-wide associational study of infectious diseases. PLoS One 2010;5:e9535. [Crossref] [PubMed]
- Lu K, Yang K, Niyongabo E, et al. Integrated network analysis of symptom clusters across disease conditions. J Biomed Inform 2020;107:103482. [Crossref] [PubMed]
- Chan SY, Loscalzo J. The emerging paradigm of network medicine in the study of human disease. Circ Res 2012;111:359-74. [Crossref] [PubMed]
- Moreau E. Literature-based discovery: addressing the issue of the subpar evaluation methodology. Bioinformatics 2023;39:btad090. [Crossref] [PubMed]
- Lang D, Chien GT. wordcloud2: Create Word Cloud by ‘htmlwidget’. 2018. doi:
10.32614/CRAN.package.wordcloud2 10.32614/CRAN.package.wordcloud2 - Vanoni M, Palumbo P. Editorial overview: Network analysis and experimental models for the understanding of multifactorial human diseases. Curr Opin Biotechnol 2020;63:vi-viii. [Crossref] [PubMed]
- Herregods TV, Bredenoord AJ, Smout AJ. Pathophysiology of gastroesophageal reflux disease: new understanding in a new era. Neurogastroenterol Motil 2015;27:1202-13. [Crossref] [PubMed]
- Layne SJ, Lorsch ZS, Patel A. Novel Diagnostic Techniques in the Evaluation of Gastroesophageal Reflux Disease (GERD). Dig Dis Sci 2023;68:2226-36. [Crossref] [PubMed]
- Zhao Y, Ma T, Zou D. Identification of Unique Transcriptomic Signatures and Hub Genes Through RNA Sequencing and Integrated WGCNA and PPI Network Analysis in Nonerosive Reflux Disease. J Inflamm Res 2021;14:6143-56. [Crossref] [PubMed]
- Böhmer AC, Schumacher J. Insights into the genetics of gastroesophageal reflux disease (GERD) and GERD-related disorders. Neurogastroenterol Motil 2017; [Crossref] [PubMed]
- Tack J, Pandolfino JE. Pathophysiology of Gastroesophageal Reflux Disease. Gastroenterology 2018;154:277-88. [Crossref] [PubMed]
- Ong JS, An J, Han X, et al. Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett’s oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut 2022;71:1053-61. [Crossref] [PubMed]
- Fairlie T, Shah A, Talley NJ, et al. Overlap of disorders of gut-brain interaction: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2023;8:646-59. [Crossref] [PubMed]
- Zhang T, Zhang B, Ma X, et al. Research trends in the field of the gut-brain interaction: Functional dyspepsia in the spotlight - An integrated bibliometric and science mapping approach. Front Neurosci 2023;17:1109510. [Crossref] [PubMed]
- Fuertes MCM, Jose BMD, Nem Singh MAA, et al. The moderating effects of information overload and academic procrastination on the information avoidance behavior among Filipino undergraduate thesis writers. Journal of Librarianship and Information Science 2020;52:694-712.
- Gong HC, Deng SH, Cao GH. Analysis of Potential Causes of Information Avoidance Behavior in Knowledge Exchange of Scientific Research. Journal of Intelligence 2022;41:189-99.
- Zhang J, Hu X, Wu D, et al. Exploring the Influence Mechanism of Chinese Young Researchers’ Academic Information Avoidance Behavior. The Journal of Academic Librarianship 2023;49:102649. [Crossref]
- Yuan X, Wang C. Research on the formation mechanism of information cocoon and individual differences among researchers based on information ecology theory. Front Psychol 2022;13:1055798. [Crossref] [PubMed]
- Choksi Y, Lal P, Slaughter JC, et al. Esophageal Mucosal Impedance Patterns Discriminate Patients With Eosinophilic Esophagitis From Patients With GERD. Clin Gastroenterol Hepatol 2018;16:664-671.e1. [Crossref] [PubMed]
- Miwa H, Kondo T, Oshima T, et al. Esophageal sensation and esophageal hypersensitivity - overview from bench to bedside. J Neurogastroenterol Motil 2010;16:353-62. [Crossref] [PubMed]
- Broekaert D, Fischler B, Sifrim D, et al. Influence of citalopram, a selective serotonin reuptake inhibitor, on oesophageal hypersensitivity: a double-blind, placebo-controlled study. Aliment Pharmacol Ther 2006;23:365-70. [Crossref] [PubMed]
- Jiang D, Zhuang Q, Jia X, et al. Current complementary and alternative therapy forgastroesophageal reflux disease. Gastroenterol Rep (Oxf) 2023;11:goad057. [Crossref] [PubMed]
- Dunbar KB, Agoston AT, Odze RD, et al. Association of Acute Gastroesophageal Reflux Disease With Esophageal Histologic Changes. JAMA 2016;315:2104-12. [Crossref] [PubMed]
- Durazzo M, Lupi G, Cicerchia F, et al. Extra-Esophageal Presentation of Gastroesophageal Reflux Disease: 2020 Update. J Clin Med 2020;9:2559. [Crossref] [PubMed]
- Ho CE, Goh YL, Zhao XX, et al. GERD: An Alternative Perspective. Psychosomatics 2016;57:142-51. [Crossref] [PubMed]
- Zhang B, Hu Y, Shi X, et al. Integrative Effects and Vagal Mechanisms of Transcutaneous Electrical Acustimulation on Gastroesophageal Motility in Patients With Gastroesophageal Reflux Disease. Am J Gastroenterol 2021;116:1495-505. [Crossref] [PubMed]
- Hershcovici T, Fass R. Pharmacological management of GERD: where does it stand now? Trends Pharmacol Sci 2011;32:258-64. [Crossref] [PubMed]
- Kiemer L, Cesareni G. Comparative interactomics: comparing apples and pears? Trends Biotechnol 2007;25:448-54. [Crossref] [PubMed]
- Futschik ME, Chaurasia G, Herzel H. Comparison of human protein-protein interaction maps. Bioinformatics 2007;23:605-11. [Crossref] [PubMed]
Cite this article as: Chen J, Chen S, Zhuang Q, Chen F, Tan N, Xiao Y. Topics and trends in gastroesophageal reflux disease research over the past 60 years: a text mining and network analysis. Transl Gastroenterol Hepatol 2025;10:29.


