
PIM1 inhibitor SMI-4a attenuated concanavalin A-induced acute hepatitis through suppressing inflammatory responses
Highlight box
Key findings
• Serine/threonine kinase 1 (PIM1) inhibitor SMI-4a attenuated concanavalin A (ConA)-induced acute hepatitis through the reduction of cleaved caspase-3 activation and phosphorylated p65 activity.
What is known and what is new?
• PIM1 inhibitor SMI-4a displays potential therapeutic effects, including inhibiting inflammation and apoptosis.
• PIM1 inhibitor SMI-4a exerts a protective effect against ConA-induced acute hepatitis.
What is the implication, and what should change now?
• The potential clinical therapeutic value and the specific molecular mechanism of PIM1 inhibitor SMI-4a might be explored in future studies.
Introduction
Hepatitis, a condition marked by liver inflammation, has shown a concerning increase worldwide over the past few decades, posing a significant risk to human health. Despite extensive research into the underlying mechanisms and the development of various treatments, a definitive cure for the disease remains elusive. Acute hepatitis cases often involve severe innate inflammation, leading to cell death and the potential development of liver failure. A crucial factor in this process is the involvement of activated macrophages, which infiltrate the liver parenchyma and contribute to its damage.
Serine/threonine kinase 1 (PIM1) is a type of serine or threonine kinase that plays a significant role in diverse cellular processes, such as proliferation, differentiation, and apoptosis (1,2). Although PIM1 was initially identified as a proto-oncogene in hematologic neoplasms, it has been shown to be involved in inflammation-related signaling pathways (3-5). In recent studies, inhibition of PIM1 has been observed to exhibit protective impacts on the progression of airway hyperresponsiveness and inflammation in mice that have been sensitized and challenged with allergens (6,7). Moreover, PIM1 kinase has been shown to play a crucial role in the regulation of pro-inflammatory mediators in fetal membranes, indicating its potential as an anti-inflammatory target (8-10).
Based on the preceding literature review and research outcomes, our hypothesis is that PIM1 potentially plays a role in the pathogenic progression of acute hepatitis induced by concanavalin A (ConA) (11). The purpose of this study is through studying PIM1 kinase of ConA-induced acute hepatitis to explore the influence of the potential mechanism of in vivo. We present this article in accordance with the ARRIVE reporting checklist (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-93/rc).
Methods
Regents
ConA was purchased from Sigma-Aldrich (Sigma-Aldrich Corporation, St. Louis; MO, USA). Selleck Corporation provided the PIM1 targeted inhibitor SMI-4a (CAS:438190-29-5, purity 100%, chemical formula C11H6F3NO2S). For mouse treatment, SMI-4a was dissolved in vegetable oil, while dimethyl sulfoxide (DMSO) was used for cell culture purposes. The antibodies used in this study include phosphorylated p65 (p-p65; from CST #3033, Cell Signaling Technology, Danvers, MA, USA) and cleaved caspase-3 (c-caspase-3; from CST #9961, Cell Signaling Technology).
Animals
Male C57/BL six mice (8 weeks old, 23±2 g) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). Mice were housed in a clean room at 24±2 °C with a 12-h light/dark cycle and had free access to food and water. Animal experiments were approved by the Clinical Center Laboratory Animal Welfare & Ethics Committee of Shanghai General Hospital, Shanghai Jiao Tong University (Ethical Batch No. 2020AWS0032). The national guideline GB/T 34791-2017 was followed for the care and use of animals.
Drug administration
Acute mouse hepatitis model was established by caudal vein method ConA with 20, 15, or 12 mg/kg (12). The mortality rate varies with different concentrations. We selected the appropriate concentration ConA (12 mg/kg). SMI-4a was given 60 mg/kg before injection.
Survival experiment
In survival experiments, 30 mice were then randomly assigned to one of four groups, the sham group (n=6), the ConA (20 mg/kg) group (n=8), the ConA (15 mg/kg) group (n=8) and the ConA (12 mg/kg) group (n=8). The survival was recorded every 3 h for 24 h.
In the SMI-4a inhibitor survival trial, we choose ConA with 12 mg/kg. Thirty mice were then randomly assigned to one of four groups: the sham group (n=6), the ConA12h group (n=8), the DMSO-ConA12h group (n=8), the SMI-4a (60 mg/kg)-ConA12h group (n=8). Then the survival was also recorded every 3 h for 24 h.
Experimental group design
Twenty-four mice were then randomly assigned to one of four groups: the sham group (n=6), the ConA3h group (n=6), the ConA12h group (n=6), and the ConA24h group (n=6). Liver tissues and serum were collected at different time points (0, 3, 12, and 24 h).
Twenty-four mice were then randomly assigned to one of four groups: the sham group (n=6), the ConA12h group (n=6), the DMSO-ConA12h group (n=6), and the SMI-4a-ConA12h group (n=6). The administration of SMI-4a (60 mg/kg) dissolved in vegetable oil was performed via intragastric delivery, 24 h prior to the caudal intravenous injection treatment of ConA. Phosphate-buffered saline (PBS) was used as control in two groups and DMSO was used as control in one group. Mice were euthanized by carbon dioxide inhalation at designated intervals.
Cells experimental design
Macrophage (RAW 264.7) models were given ConA (400 µg/mL) 12 h and SMI-4a was given 80 µM 24 h before ConA given to establish cell models, the dosage about SMI-4a was directly referred to the experiment (13). Cells are collected after 12 h. The cells are divided into four groups: the control group, the ConA12h group, the DMSO-ConA12h group, and the SMI-4a-ConA12h group.
Biochemical analysis
At each time point (0, 3, 12, and 24 h) following ConA injection, we euthanized six mice randomly chosen from each group. We obtained liver tissues from the mice, which were subsequently stored at −80 °C, and collected cardiac puncture blood samples after centrifugation, which were stored at 4 °C.
Histology evaluation
Liver tissue samples were collected at 0, 3, 12, and 24 h after ConA caudal vein injection. Afterwards, the upper right lobe was immersed in 4% paraformaldehyde for fixation, followed by dehydration using ethanol, and finally embedded in paraffin. The liver tissue was sliced into sections measuring 4–5 µm in thickness and subsequently underwent staining with hematoxylin-eosin (HE). An optical microscope was employed to analyze and evaluate the pathological variations within the four groups. The scoring procedure was carried out independently by two skilled pathologists, following the previously established protocol. Pathological score: 1, inactivity; 2, mild active, punctate necrosis; 3, mild activity, mild lobular necrosis; 4, moderate activity, moderate lobular necrosis; 5, severe activity, severe lobular necrosis.
Serum aminotransferase assessment
Following a 4–5 h storage at 4 °C, separation of sera from cardiac puncture blood collection was carried out through centrifugation at 2,000 rpm for 10 minutes. To assess hepatocellular injury, measurement of serum levels for alanine aminotransferase (ALT) and aspartate aminotransferase (AST) was performed using microplate test kits specific for ALT and AST, following the guidelines provided by the manufacturer. The ALT and AST microplate test kits were purchased from Nanjing Jiancheng Bioengineering Institute (Jiancheng Biotech, Nanjing, China).
Serum cytokine assays
To measure serum proinflammatory cytokines tumor necrosis factor (TNF)-α, interleukin (IL)-6, and IL-1β, enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems, Minneapolis, MN, USA) were utilized in accordance with the protocols provided by the manufacturers.
Flow cytometry (FCM)
The infiltration of neutrophils, T cells, and macrophages was detected by FCM. FITC-CD11b (eBioscienc, 11-0112-82; San Diego, CA, USA) and PE-Ly-6G (BD, 551461; Franklin Lakes, NJ, USA) were used to label neutrophils, FITC and F4/80 (BioLegend, MF48005; San Diego, CA, USA) antibodies were used to label macrophages. CD4-APC (eBioscience, 17-0041-81) and CD8-FITC (eBioscienc, 11-0086-42) were used to T cells.
Western blotting analysis
Following the freezing of liver tissues in liquid nitrogen, the tissues were quickly disrupted using radio-immunoprecipitation assay (RIPA) lysis buffer that contained various components such as phenylmethanesulfonyl fluoride (PMSF), aprotinin, sodium orthovanadate, and sodium fluoride (purchased from Sigma-Aldrich). To determine the protein concentration, the bicinchoninic acid (BCA) method was employed. To prepare the samples for analysis, equivalent quantities of total protein (30–120 g) were subjected to boiling with 5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer. The treated samples were then examined using SDS-PAGE as per the standardized protocols. In order to prevent nonspecific binding, 5% nonfat milk dissolved in PBS was applied for a period of 2 h. The blots were subsequently incubated overnight at 4 °C with rabbit antibodies that targeted either mouse α-tubulin (1:1,000), mouse p-p65 (1:1,000), or rabbit c-caspase-3 (1:1,000), all prepared in 5% milk. The cytoplasmic proteins were normalized using α-tubulin as an internal reference, while nuclear proteins were normalized using lamin-A. After washing the membranes three times with PBS with Tween 20 (PBST), a secondary antibody (either goat anti-mouse or anti-rabbit) was applied at a dilution of 1:2,000 in PBST for incubation at 37 °C for 60 minutes. Lastly, the membranes were washed three times with PBST for 5 minutes each, and the proteins were detected using the Odyssey two-color infrared laser imaging system, which utilizes fluorescence detection. The antibodies employed in this research include p-p65 (from CST #3033) and c-caspase-3 (from CST #9961).
Quantitative real-time polymerase chain reaction (qRT-PCR) assay
Using a commercial kit (miRNeasy Mini Kit, Qiagen, Hilden, Germany), liver tissue was subjected to various time intervals after ConA administration in order to extract total RNA. The RNA quality was assessed by analyzing the A260/A280 ratio, followed by the synthesis of complementary DNA (cDNA) using reverse transcriptase (Takara, Kusatsu, Japan). For each sample, triplicate preparations were made, resulting in a total reaction volume of 20 µL. The reaction consisted of 250 nM forward and reverse primers, 10 µL SYBR Green (Takara), ROX Reference Dye II, and 20 ng cDNA. RT-PCR was performed using the QuantStudio 3 system. To account for variability in expression levels, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as the internal control for normalization (14). The PCR primers used in the experiment are shown in Table 1.
Table 1
| Primer | Sequence |
|---|---|
| PIM1 primer | Forward GCTCGGTCTACTCTGGCATC |
| Reverse CCGAGCTCACCTTCTTCAAC | |
| GAPDH primer | Forward AACTTTGGCATTGTGGAAGG |
| Reverse GGATGCAGGGATGATGTTCT | |
| TNF-α primer | Forward CACCACCATCAAGGACTCAAAT |
| Reverse TCAGGGAAGAATCTGGAAAGGT | |
| IL-6 primer | Forward GGCCCTTGCTTTCTCTTCG |
| Reverse ATAATAAAGTTTTGATTATGT | |
| IL-1β primer | Forward GAAAGCTCTCCACCTCAATG |
| Reverse GCCGTCTTTCATTACACAGG |
PCR, polymerase chain reaction; PIM1, serine/threonine kinase 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IL, interleukin; TNF, tumor necrosis factor.
Statistical analysis
Data were represented as the average ± standard deviation. Analyzing the statistical data was conducted with the assistance of the SPSS Statistics version 17.0 Program (SPSS Inc., Chicago, IL, USA). Prism 7.0 (GraphPad Software, San Diego, CA, USA) was employed for creating graphical representations. The survival data were assessed through the utilization of Kaplan-Meier plots and log-rank tests. Comparisons among the groups were performed via one-way analysis of variance (ANOVA), followed by Tukey’s multiple comparison test. Statistical significance was defined as P<0.05.
Results
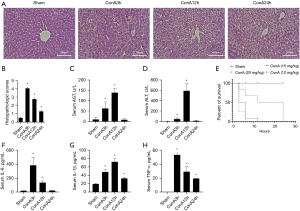
The severity of acute hepatitis is related to the concentration and duration of ConA
The study found that the most severe liver injury occurred at 3 and 12 h after ConA injection, as observed by HE staining of liver tissue in Figure 1A and pathological scoring in Figure 1B. The inflammation of liver tissue was increased significantly, accompanied by massive necrosis of hepatic lobules in 3 and 12 h. Serum AST and ALT levels were highest at 12 h after ConA injection in Figure 1C,1D. The survival rate of mice significantly decreased with increasing concentrations of ConA in Figure 1E. Therefore, ConA 12 mg/kg is the most appropriate experimental dose. In addition, the expression of serum inflammatory cytokines (IL-6, IL-1β, and TNF-α) significantly increased at 3 and 12 h after ConA injection in Figure 1F-H.
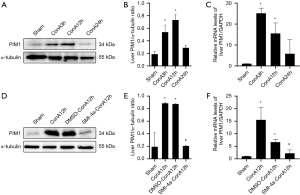
PIM1 inhibitor SMI-4a can inhibit PIM1 expression in acute hepatitis
In the acute hepatitis model, the expression of PIM1 increased significantly at 3 and 12 h, while PIM1 protein expression was highest at 12 h after ConA injection, as shown in Figure 2A,2B. PIM1 messenger RNA (mRNA) expression also increased significantly at 3 and 12 h, then at 12 h the increase of PIM1 mRNA expression was the most obvious after ConA injection, as shown in Figure 2C. When treated with the PIM1 inhibitor, SMI-4a, PIM1 protein expression was significantly inhibited at 12 h, as shown in Figure 2D,2E. Similarly, PIM1 mRNA expression was also inhibited by SMI-4a treatment at 12 h, as shown in Figure 2F. These results suggest that the ConA model can cause PIM1 to rise, and the inhibitor SMI-4a can effectively inhibit PIM1 expression in the acute hepatitis model.
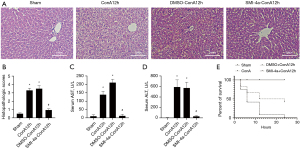
PIM1 inhibitor SMI-4a improves liver injury in ConA-induced acute hepatitis
In the acute hepatitis model, treatment with PIM1 inhibitor SMI-4a improved liver injury observed 12 h after caudal intravenous injection of ConA (12 mg/kg) as shown in HE staining in Figure 3A. After the addition of SMI-4a inhibitors, the inflammatory response of liver tissue was obviously limited, and the necrosis of liver lobular structure was significantly reduced. Pathological score also indicated improvement in liver injury after PIM1 inhibitor treatment as shown in Figure 3B. Serum AST and ALT levels were reduced after treatment with PIM1 inhibitor at 12 h as shown in Figure 3C,3D. The survival rate of mice treated with ConA (12 mg/kg) and PIM1 inhibitor was significantly increased compared to the ConA group in Figure 3E.
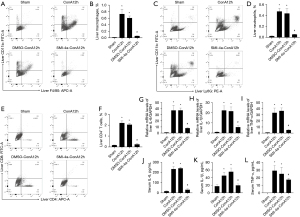
PIM1 inhibitor SMI-4a improves inflammation in ConA-induced acute hepatitis
In this study, we found that SMI-4a can inhibit the count of macrophages in liver tissue as shown in Figure 4A,4B. Then, we observed that treatment with SMI-4a can restrain the count of neutrophils in liver tissue as shown in Figure 4C,4D. We found that SMI-4a can inhibit the count of CD4+T cells in liver tissue after SMI-4a treatment 12 h as shown in Figure 4E,4F. IL-6, IL-1β, and TNF-α mRNA expression were also inhibited by SMI-4a treatment at 12 h, as shown in Figure 4G-4I. Furthermore, the expression of inflammatory cytokines (IL-6, IL-1β, and TNF-α) in serum was significantly decreased comparison with ConA injection after PIM1 inhibitor treatment in Figure 4J-4L. Therefore, SMI-4a can inhibit the production of immune cells and inhibit inflammatory factors in liver tissues and serum.
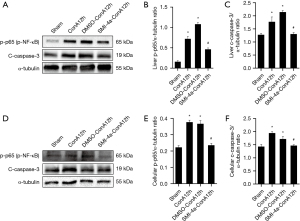
SMI-4a protects against liver damage through inhibiting p65 nuclear factor (NF)-κB phosphorylation and caspase-3 cleaving
In this investigation, we initially validated that the molecule SMI-4a possesses the capability to suppress the manifestation of the PIM1 protein. Subsequently, we observed that treatment with SMI-4a down-regulated the levels of p-p65 and c-caspase-3 in the acute hepatitis model in Figure 5A-5C. Similarly, in the RAW 264.7 model, treatment with SMI-4a also down-regulated the levels of p-p65 and c-caspase-3, as shown in Figure 5D-5F.
Discussion
In this study, we used ConA to induce acute liver injury (15,16). There is no special treatment for acute autoimmune hepatitis in clinical practice, and it is treated by immunosuppressants and hormones. Our study could provide a drug that could be considered for autoimmune acute hepatitis. In order to provide new treatment ideas, we found that different concentrations of ConA not only showed different survival rates, but also had different severity of liver damage and inflammatory response at different time points. Meanwhile, PIM1 expression was the most significant at 12 h time point. We found that SMI-4a can reduce the number of macrophages, neutrophils, and T cells, and it is very noticeable in macrophages. The PIM1 inhibitor SMI-4a inhibits the expression of PIM1 and also inhibits liver damage and inflammatory responses, while acting on macrophages (17,18). ConA-induced liver injury after SMI-4a pretreatment has been shown to reduce inflammatory response and liver injury through reduced expression of p-p65 and c-caspase-3, both in vivo and in vitro.
Previous literature found that different concentrations of ConA had different effects on liver injury (19,20). Therefore, we observed the survival rate of mice through different ConA concentration gradients, and selected the most appropriate concentration of 12 mg/kg for the experiment (21). It was found in the experiment that the onset of acute hepatitis reaction was acute at 3 h. AST and ALT had increased significantly, and reached a peak at 12 h. HE staining showed that the liver damage at 12 h was very serious. At the same time, we found that the expression level of PIM1 increased most significantly at 12 h. Therefore, we believe that inhibition of PIM1 expression is needed to alleviate acute liver injury (22-24). In vivo experiments, we found that the pro-inflammatory factors TNF-α, IL-6, and IL-1β showed an increasing trend at 3 and 12 h while a decreasing trend at 24 h. At the same time, we found that oral SMI-4a preconditioning can reduce PIM1 levels in liver tissue in vivo. These findings suggest that a number of complex pathway inflammatory and compensatory mechanisms are involved, which rely on PIM1 activation in response to inflammatory stimuli in the body.
As a kind of serine/threonine kinase, PIM1 role in different types of tumor infiltrating has attracted much attention (25,26). Interestingly, recent studies have reported an association between PIM1 and inflammatory diseases (27-29). In our study, we found that PIM1 mRNA and protein in the liver tissue of acute hepatitis induced by ConA. PIM1 may participate in the development of ConA-induced acute hepatitis process (30). In addition, preconditioning with PIM1-specific inhibitor SMI-4a improved the prognosis of ConA-induced acute hepatitis (31). In vivo experiments, we found that the pro-inflammatory factors TNF-α, IL-6, and IL-1β can be inhibited by SMI-4a.
The NF-κB is an inducible transcription factor that plays a role in regulating the expression level of various proteins involved in cell survival and immune response (31,32). We detected the role of SMI-4a in phosphorylation of p65 and changes in phosphorylation of p65 in liver tissue or macrophage cell lines. The expression of p65 phosphorylation was up-regulated by ConA. SMI-4a treatment reduced the phosphorylation level of p65. At the same time, the role of SMI-4a in caspase-3 shearing and the change of caspase-3 shearing in liver tissue or macrophage cell line. The expression of caspase-3 splicing was up-regulated by ConA. SMI-4a treatment reduced caspase-3 shear levels. These findings may explain the decrease in cytokine production.
In summary, we found that macrophages are involved in ConA-induced acute hepatitis. SMI-4a significantly inhibits PIM1 and the release of inflammatory cytokines, acting by inhibiting the phosphorylation of p65 and the shear of caspase-3. Our experimental results show that inhibiting PIM1 expression is of great significance to relieve acute hepatitis.
Conclusions
The PIM1 inhibitor SMI-4a inhibited the release of inflammatory cytokines by inhibiting p-p65 activation and c-caspase-3 activity. The PIM1 inhibitor SMI-4a developed a good protective effect on the experimental mouse model of ConA-induced acute hepatitis.
Acknowledgments
Funding: This study was funded by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-93/rc
Data Sharing Statement: Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-93/dss
Peer Review File: Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-93/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-93/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were approved by the Clinical Center Laboratory Animal Welfare & Ethics Committee of Shanghai General Hospital, Shanghai Jiao Tong University (Ethical Batch No. 2020AWS0032). The national guideline GB/T 34791-2017 was followed for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shin YS, Takeda K, Shiraishi Y, et al. Inhibition of Pim1 kinase activation attenuates allergen-induced airway hyperresponsiveness and inflammation. Am J Respir Cell Mol Biol 2012;46:488-97. [Crossref] [PubMed]
- de Vries M, Heijink IH, Gras R, et al. Pim1 kinase protects airway epithelial cells from cigarette smoke-induced damage and airway inflammation. Am J Physiol Lung Cell Mol Physiol 2014;307:L240-51. [Crossref] [PubMed]
- Lim R, Barker G, Lappas M. Inhibition of PIM1 kinase attenuates inflammation-induced pro-labour mediators in human foetal membranes in vitro. Mol Hum Reprod 2017;23:428-40. [Crossref] [PubMed]
- Yoon SB, Hong H, Lim HJ, et al. A novel IRAK4/PIM1 inhibitor ameliorates rheumatoid arthritis and lymphoid malignancy by blocking the TLR/MYD88-mediated NF-κB pathway. Acta Pharm Sin B 2023;13:1093-109. [Crossref] [PubMed]
- Li Z, Lin F, Zhuo C, et al. PIM1 kinase phosphorylates the human transcription factor FOXP3 at serine 422 to negatively regulate its activity under inflammation. J Biol Chem 2014;289:26872-81. [Crossref] [PubMed]
- Wang J, Cao Y, Liu Y, et al. PIM1 inhibitor SMI-4a attenuated lipopolysaccharide-induced acute lung injury through suppressing macrophage inflammatory responses via modulating p65 phosphorylation. Int Immunopharmacol 2019;73:568-74. [Crossref] [PubMed]
- Cao Y, Chen X, Liu Y, et al. PIM1 inhibition attenuated endotoxin-induced acute lung injury through modulating ELK3/ICAM1 axis on pulmonary microvascular endothelial cells. Inflamm Res 2021;70:89-98. [Crossref] [PubMed]
- Liu Y, Shang Y, Yan Z, et al. Pim1 kinase positively regulates myoblast behaviors and skeletal muscle regeneration. Cell Death Dis 2019;10:773. [Crossref] [PubMed]
- Zhang X, Zou Y, Liu Y, et al. Inhibition of PIM1 kinase attenuates bleomycin-induced pulmonary fibrosis in mice by modulating the ZEB1/E-cadherin pathway in alveolar epithelial cells. Mol Immunol 2020;125:15-22. [Crossref] [PubMed]
- Ren W, Wang X, Yang M, et al. Distinct clinical and genetic features of hepatitis B virus-associated follicular lymphoma in Chinese patients. Blood Adv 2022;6:2731-44. [Crossref] [PubMed]
- Luo Y, Zhu J, Zhao F, et al. PIM1 attenuates renal ischemia-reperfusion injury by inhibiting ASK1-JNK/P38. Int Immunopharmacol 2023;114:109563. [Crossref] [PubMed]
- Park C, Min S, Park EM, et al. Pim Kinase Interacts with Nonstructural 5A Protein and Regulates Hepatitis C Virus Entry. J Virol 2015;89:10073-86. [Crossref] [PubMed]
- Yao X, Jin G, Liu D, et al. Inducible nitric oxide synthase regulates macrophage polarization via the MAPK signals in concanavalin A-induced hepatitis. Immun Inflamm Dis 2022;10:e643. [Crossref] [PubMed]
- Narlik-Grassow M, Blanco-Aparicio C, Cecilia Y, et al. Conditional transgenic expression of PIM1 kinase in prostate induces inflammation-dependent neoplasia. PLoS One 2013;8:e60277. [Crossref] [PubMed]
- Shen YM, Zhao Y, Zeng Y, et al. Inhibition of Pim-1 kinase ameliorates dextran sodium sulfate-induced colitis in mice. Dig Dis Sci 2012;57:1822-31. [Crossref] [PubMed]
- Heymann F, Hamesch K, Weiskirchen R, et al. The concanavalin A model of acute hepatitis in mice. Lab Anim 2015;49:12-20. [Crossref] [PubMed]
- Liu C, Gao S, Qu Z, et al. NCPP treatment alleviates ConA-induced hepatitis via reducing CD4+T activation and NO production. Immunopharmacol Immunotoxicol 2012;34:962-7. [Crossref] [PubMed]
- Wang Y, Guo X, Jiao G, et al. Splenectomy Promotes Macrophage Polarization in a Mouse Model of Concanavalin A- (ConA-) Induced Liver Fibrosis. Biomed Res Int 2019;2019:5756189. [Crossref] [PubMed]
- Zemskova MY, Song JH, Cen B, et al. Regulation of prostate stromal fibroblasts by the PIM1 protein kinase. Cell Signal 2015;27:135-46. [Crossref] [PubMed]
- Vries Md, Bedke N, Smithers NP, et al. Inhibition of Pim1 kinase, new therapeutic approach in virus-induced asthma exacerbations. Eur Respir J 2016;47:783-91. [Crossref] [PubMed]
- Wang M, Okamoto M, Domenico J, et al. Inhibition of Pim1 kinase prevents peanut allergy by enhancing Runx3 expression and suppressing T(H)2 and T(H)17 T-cell differentiation. J Allergy Clin Immunol 2012;130:932-44.e12. [Crossref] [PubMed]
- de Vries M, Smithers NP, Howarth PH, et al. Inhibition of Pim1 kinase reduces viral replication in primary bronchial epithelial cells. Eur Respir J 2015;45:1745-8. [Crossref] [PubMed]
- Moore G, Lightner C, Elbai S, et al. Co-Targeting PIM Kinase and PI3K/mTOR in NSCLC. Cancers (Basel) 2021;13:2139. [Crossref] [PubMed]
- Fischer KM, Cottage CT, Konstandin MH, et al. Pim-1 kinase inhibits pathological injury by promoting cardioprotective signaling. J Mol Cell Cardiol 2011;51:554-8. [Crossref] [PubMed]
- Zou Y, Cao Y, Liu Y, et al. The role of dorsal root ganglia PIM1 in peripheral nerve injury-induced neuropathic pain. Neurosci Lett 2019;709:134375. [Crossref] [PubMed]
- Liu JY, Wang KX, Huang LY, et al. Expression and role of Pim1 in cultured cortical neurons with oxygen-glucose deprivation/reoxygen injury. Zhongguo Dang Dai Er Ke Za Zhi 2020;22:512-8. [PubMed]
- Jiménez-García MP, Lucena-Cacace A, Robles-Frías MJ, et al. Inflammation and stem markers association to PIM1/PIM2 kinase-induced tumors in breast and uterus. Oncotarget 2017;8:58872-86. [Crossref] [PubMed]
- de Vries M, Hesse L, Jonker MR, et al. Pim1 kinase activity preserves airway epithelial integrity upon house dust mite exposure. Am J Physiol Lung Cell Mol Physiol 2015;309:L1344-53. [Crossref] [PubMed]
- Luan J, Zhang X, Wang S, et al. NOD-Like Receptor Protein 3 Inflammasome-Dependent IL-1β Accelerated ConA-Induced Hepatitis. Front Immunol 2018;9:758. [Crossref] [PubMed]
- Li N, Wu JJ, Qi M, et al. CP-25 exerts a protective effect against ConA-induced hepatitis via regulating inflammation and immune response. Front Pharmacol 2022;13:1041671. [Crossref] [PubMed]
- Wang K, Wu W, Jiang X, et al. Multi-Omics Analysis Reveals the Protection of Gasdermin D in Concanavalin A-Induced Autoimmune Hepatitis. Microbiol Spectr 2022;10:e0171722. [Crossref] [PubMed]
- Fang X, Wang R, Ma J, et al. Ameliorated ConA-induced hepatitis in the absence of PKC-theta. PLoS One 2012;7:e31174. [Crossref] [PubMed]
Cite this article as: Wu X, Chen Y, Jiang M, Guo L. PIM1 inhibitor SMI-4a attenuated concanavalin A-induced acute hepatitis through suppressing inflammatory responses. Transl Gastroenterol Hepatol 2024;9:14.


