
Esophageal carcinoma with SMARCA4 mutation: a narrative review for this rare entity
Introduction
Switch/sucrose nonfermenting (SWI/SNF)-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4 (SMARCA4) or BRG1 protein is a nuclear protein involved in chromatin remodeling that is encoded by the SMARCA4 gene (1). As part of the large ATP-dependent chromatin remodeling complex SWI/SNF, the SMARCA4/BRG1 protein is required for the transcriptional activation of genes normally repressed by chromatin through epigenetic changes and is necessary for normal cellular growth and proliferation (2,3). Recent studies have also found that epigenetic modifications play critical roles in cancer development, with 20% of cancer patients harboring SWI/SNF mutations (4,5). SMARCA4 mutations have been reported in multiple malignancies, affecting the lungs, uterus, skin, and head and neck (6-12).
Numerous studies have shown that SMARCA4 mutations mainly contribute to carcinogenesis in two aspects. First, SMARCA4/BRG1 proteins were direct regulators for transcriptional activation of genes related to carcinogenesis. For example, loss of SMARCA4/BRG1 has been shown to cause overexpression of oncoprotein MYC in lung cancer, which enabled cancer cells to sustain undifferentiated gene expression programs and prevented its response to environmental stimuli (13). In breast cancer cells, loss of SMARCA4/BRG1 reduced its binding to BRCA1 protein, which disrupted the tumor suppresser function of BRCA1 (14). Deletion of SMARCA4/BRG1 also caused increased expression of CD44 in cell cultures and decreased CDK4/6 kinase activity in ovarian cancer cells, resulting in cell cycle progressing, increased tumor growth, and metastasis (15,16). Secondly, SMARCA4/BRG1 protein played important roles in DNA processing as an essential component of chromatin remodeling complex in addition to its transcription regulating roles, which included regulating and promoting DNA repair by repositioning nucleosomes and recruiting other DNA repair proteins (17-19). In human lung cancer and breast cancer, the absence of SMARCA4/BRG1 protein has been demonstrated to be a major cause of genome instability (20-22).
Esophageal carcinoma with SMARCA4 mutation is a rare variant of malignant esophageal epithelial neoplasm, which is characterized by SMARCA4/BRG1 protein loss on immunohistochemistry or SMARCA4 gene alteration on sequencing. Kilic et al. first reported a 70-year-old patient who was presented with an undifferentiated gastroesophageal junction (GEJ) carcinoma with liver metastasis (23). This disease can be considered a variant of undifferentiated esophageal carcinoma according to the World Health Organization classification, which typically lacks the overly common features of squamous, glandular, or neuroendocrine differentiation (24). However, only a few case series and case reports of esophageal carcinoma with SMARCA4 mutations have been published in the English literature (23,25-32). To the best of our knowledge, there are no previously published comprehensive review articles for esophageal carcinoma with SMARCA4 mutations covering its epidemiological, clinical, pathological, and molecular features. The rarity of the disease poses significant diagnostic challenges for surgical pathologists and can potentially lead to delayed or suboptimal patient care. Herein, we reviewed the available literature on esophageal carcinoma with SMARCA4 mutations to discuss its epidemiological, clinical, pathological, and molecular features with diagnostic challenges, as well as provide an overview of its treatment and prognosis. We present this article in accordance with the Narrative Review reporting checklist (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-84/rc).
Methods
A search was conducted in the PubMed, Scopus, Ovid, and Google Scholar databases using selected keywords (Table 1). The search was originally conducted on August 1, 2023, and updated on January 25, 2024. Two independent researchers reviewed each article to ensure that it met the inclusion and exclusion criteria. All duplicates were excluded from the analysis. The references of all searched articles were cross-examined to avoid missing other relevant studies.
Table 1
| Items | Specification |
|---|---|
| Date of search | August 1, 2023, and updated on January 25, 2024 |
| Databases and other sources searched | PubMed, Scopus, Ovid, and Google Scholar |
| Search terms used | Esophagus AND SMARCA4 AND carcinoma |
| Timeframe | From inception till January 25, 2024 |
| Inclusion and exclusion criteria | Inclusion criteria: articles published in the English literature and human participants |
| Exclusion criteria: articles published in languages other than English, and non-human subjects | |
| Selection process | Two authors separately conducted the initial database search using the abovementioned search terms. Final search result was achieved by consensus |
SMARCA4, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4; SWI/SNF, switch/sucrose nonfermenting.
Articles related to human cancer that provided descriptions of the clinical, pathological, and molecular features and were published in English were selected. Esophageal carcinoma involving the GEJ is mostly staged as esophageal carcinoma in surgical pathology practice. Studies regarding this subject were also included.
Articles published in languages other than English, articles on non-human cancer, non-clinical basic research articles, articles on esophageal carcinoma with other closely related mutations such as SMARCA2, and articles on carcinoma with SMRACA4 mutation from primary sites other than the esophagus or GEJ were excluded.
The clinical presentations, histopathological and molecular findings, treatment, and disease prognosis were evaluated and reported.
Search results
Nine full articles on esophageal carcinoma with SMARCA4 mutations were identified, which included case reports, case series, and retrospective case studies. Table 2 lists the nine publications and summarizes the patient characteristics, clinical presentation, associated risk factors, treatment, and follow-up information. Notably, one case reported in the study by Cui et al. (25) was previously documented in a separate case report (26); therefore, these two publications were summarized together. Similarly, one case reported in the study by Gupta et al. (28) was previously documented in a standalone case report (23); consequently, the results of these two publications were consolidated into a singular summary. The study by Neil et al. (27) was included as it contained a detailed description of the SMARCA4 gene alterations and staging information; although esophageal carcinoma and gastric carcinoma were reported together, other clinical and pathological data were not presented separately. In the study by Chang et al. (30), four GEJ carcinoma cases with SMARCA4 loss were presented which have been included in this review; the remaining 26 cases involved the gastrointestinal tracts other than the esophagus junction or GEJ and were therefore excluded. Twelve of 14 cases from the study by Horton et al. (31) showed SMARCA4 loss and were therefore included; the remaining two cases with SMARCA2 protein loss were excluded. Lastly, the study by Schallenberg et al. (32) was included because it contained epidemiological and molecular features of esophageal carcinoma with SMARCA4 mutations, despite the absence of information on clinical and pathological features.
Table 2
| Refs | Number of cases | Patient | Age (years) | Gender | Clinical presentations | Associated factors | Location | Size | Clinical stage | Treatment | Follow-up |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cui et al., 2023 (25); Cui et al., 2023 (26) | 4 | No. 1 | 68 | Female | Dysphasia and weight loss | GERD | Esophagus, distal | Large mass | IVB (liver) | Stent placement | Died, 72 days |
| No. 2 | 47 | Male | Abdominal pain | Alcohol use | Esophagus, distal | 2.5 cm | IVB (liver) | Palliative | Died, 78 days | ||
| No. 3 | 45 | Male | Dark stool | Barrett’s esophagus | Esophagus, distal | 4–5 cm | III | Chemoradiation and surgery | Died, 8 months | ||
| No. 4 | 55 | Male | Abdominal pain and weight loss | Alcohol use, Barrett’s esophagus | Esophagus, distal | Large mass | III | Chemotherapy | Died, 3 months | ||
| Neil et al., 2023 (27) | 2 | – | – | – | – | – | – | – | II | – | – |
| 5 | – | – | – | – | – | – | – | III | – | – | |
| 5 | – | – | – | – | – | – | – | IVA | – | – | |
| 16 | – | – | – | – | – | – | – | IVB | – | – | |
| Kilic et al., 2019 (23); Gupta et al., 2023 (28) | 5 | No. 1 | 53 | Male | Odynophagia and epigastric pain | GERD, smoking | Esophagus, distal | 2.1 cm | – | Chemotherapy and surgery | Died, 1 month |
| No. 2 | 48 | Male | Dysphasia and weight loss | Smoking | Esophagus, distal | 9 cm | – | Chemotherapy and surgery | Alive, 3 years | ||
| No. 3 | 79 | Female | Nausea, vomiting, and weight loss | Barrett’s esophagus | Esophagus, mid to distal | – | – | – | – | ||
| No. 4 | 70 | Male | Fatigue, poor appetite, and abdominal discomfort | – | GEJ | – | IVB (liver and lung) | Palliative | Died, 1 month | ||
| No. 5 | 70 | Male | - | Smoking and HIV+ | Esophagus, distal | 5 cm | IVB (liver) | Palliative | Alive with disease | ||
| Ahmed et al., 2022 (29) | 2 | No. 1 | 39 | Male | Fever, nausea, abdominal pain, and weight loss | Barrett’s esophagus | Esophagus, distal | Large mass | IVB (liver) | Palliative | Died, 1.5 months |
| No. 2 | 64 | Male | Chest pain | Barrett’s esophagus, GERD, smoking, and alcohol use | Esophagus, distal | 10 cm | IVB (liver) | Palliative | Died, 3.0 months | ||
| Chang et al., 2022 (30) | 4 | No. 1 | 60 | Male | – | – | GEJ | 7.0 cm | IVB (liver) | Chemoradiation and surgery | Died, 11 months |
| No. 2 | 73 | Male | – | – | GEJ | - | III | Palliative | Died, 4 months | ||
| No. 3 | 40 | Female | – | – | GEJ | 5.0 cm | IB | Surgery | Alive, 20 months | ||
| No. 4 | 85 | Male | – | – | GEJ | – | IVA | Target therapy | – | ||
| Horton et al., 2021 (31) | 12 | No. 1 | 63 | Male | – | – | GEJ | – | – | – | – |
| No. 2 | 72 | Male | – | Barrett’s esophagus | Esophagus, not specified | – | – | – | – | ||
| No. 3 | 77 | Female | – | Barrett’s esophagus | Esophagus, mid | – | – | – | – | ||
| No. 4 | 72 | Female | – | Barrett’s esophagus | Esophagus, mid | – | – | – | – | ||
| No. 5 | 70 | Male | – | Barrett’s esophagus | Esophagus, distal | – | – | – | – | ||
| No. 6 | 64 | Male | – | Barrett’s esophagus | Esophagus, not specified | – | – | – | – | ||
| No. 7 | 68 | Female | – | Barrett’s esophagus | GEJ | – | – | – | – | ||
| No. 8 | 76 | Male | – | – | Esophagus, distal | – | – | – | – | ||
| No. 9 | 63 | Male | – | Barrett’s esophagus | Esophagus, distal | – | – | – | – | ||
| No. 10 | 79 | Female | – | Barrett’s esophagus | Esophagus, distal | – | – | – | – | ||
| No. 11 | 77 | Male | – | – | Esophagus, not specified | – | – | – | – | ||
| No. 12 | 73 | Male | – | – | Esophagus, distal | – | – | – | – | ||
| Schallenberg et al., 2020 (32) | 1 | – | – | Female | – | – | Esophagus, not specified | – | – | – | – |
| 18 | – | – | Male | – | – | Esophagus, not specified | – | – | – | – |
–, data not available. SMARCA4, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4; GERD, gastroesophageal reflux disease; SWI/SNF, switch/sucrose nonfermenting; GEJ, gastroesophageal junction; HIV, human immunodeficiency virus.
In summary, 74 patients were identified and included in this review (Table 2), of whom 40 had detailed pathological and immunochemical descriptions (summarized in Table 3) and 55 had detailed descriptions of molecular features (Table 4).
Table 3
| Refs | Number of cases | Epithelioid features | Rhabdoid features | Glandular features | AE1/3 | Cam 5.2 | CK cocktail | CK Oscar | CK7 | CK20 | CDX2 | GATA3 | SALL4 | TTF1 | P40 | P63 | Synaptophysin | Chromogranin | INSM1 | CD56 | SOX10 | S100 | CD45 | CD3 | CD20 | SMARCA4 (BRG1) | SMARCB1 (INI1) | P53 | MMR | HER2 (ERBB2) | PD-L1 (CPS) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cui et al., 2023 (25); Cui et al., 2023 (26) | 4 | 4/4 | 3/4 | 2/4 | 3/3+ | NM | NM | NM | 2/2+ | 0/1+ | 1/3+ | 0/2+ | 2/2+ | 1/1+ | 1/4+ | 1/1+ | 2/3+ | 0/4+ | 0/1+ | 0/1+ | 0/1+ | 0/1+ | 0/1+ | 0/1+ | 0/1+ | 2/3 loss | 3/3 intact | 1/1 mutant | 4/4 intact | 0/2+ | 1/3+ |
| Kilic et al., 2019 (23); Gupta et al., 2023 (28) | 5 | 5/5 | 4/5 | 0/5 | 1/2+ | 2/5+ | 1/2+ | 0/1+ | 0/1+ | 0/1+ | 0/1+ | 0/1+ | NM | 0/1+ | 0/1+ | 0/2+ | 1/2+ | 0/2+ | NM | 0/3+ | 0/3+ | 0/3+ | 0/3+ | 0/4+ | 0/3+ | 5/5 loss | 5/5 intact | NM | 4/4 intact | NM | NM |
| Ahmed et al., 2022 (29) | 2 | 2/2 | 0/2 | 0/2 | NM | NM | NM | NM | NM | NM | NM | NM | 1/2+ | NM | NM | NM | M | NM | NM | NM | NM | NM | NM | NM | NM | 2/2 loss | 2/2 intact | NM | 2/2 intact | NM | NM |
| Chang et al., 2022 (30) | 4 | 4/4 | 0/4 | 0/4 | 2/4+ | NM | NM | NM | 0/1+ | NM | 0/1+ | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | 4/4 loss | 3/3 intact | NM | NM | NM | NM |
| Horton et al., 2021 (31) | 12 | NM | NM | NM | 4/11+ | 3/6+ | NM | 5/6+ | NM | NM | 2/8+ | NM | NM | NM | NM | 2/4+ | NM | NM | NM | NM | NM | 0/9+ | NM | 0/9+ | 0/9+ | 12/12 loss | 7/7 intact | 4/4 mutant | 7/7 intact | NM | NM |
| Schallenberg et al., 2020 (32) | 13 | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | 19/19 loss | NM | 6/13 | NM | 0/13+ | NM |
| Total | 40 | 15/15 | 7/15 | 2/15 | 10/20+ | 5/11+ | 1/2+ | 5/7+ | 2/4+ | 0/2+ | 3/13+ | 0/3+ | 3/4+ | 1/2+ | 1/5+ | 3/7+ | 3/5+ | 0/6+ | 0/1+ | 0/4+ | 0/4+ | 0/13+ | 0/4+ | 0/14+ | 0/13+ | 44/45 loss | 20/20 intact | 11/18 mutant | 17/17 intact | 0/15+ | 1/3+ |
| Rate | – | 100% | 47% | 13% | 50%+ | 45%+ | 50%+ | 71%+ | 50%+ | 0%+ | 23%+ | 0%+ | 75%+ | 50%+ | 20%+ | 43%+ | 60%+ | 0%+ | 0%+ | 0%+ | 0%+ | 0%+ | 0%+ | 0%+ | 0%+ | 98% loss | 100% intact | 61% mutant | 100% intact | 0%+ | 33%+ |
+, positive. SMARCA4, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4; SWI/SNF, switch/sucrose nonfermenting; MMR, mismatch repair proteins; CPS, combined positive score; NM, not mentioned.
Table 4
| Refs | Number of cases | SMARCA4 mutation | Tumor mutation burden | CCNE1 amplification | TP53 mutation | CDKN2A mutation | EGFR amplification | CTNNB1 mutation | PTPRD mutation | MET amplification | C-myc amplification | KRAS amplification | GATA6 amplification | PIK3CA amplification |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cui et al., 2023 (25); Cui et al., 2023 (26) | 4 | 4/4 | Low, 4/4 | 1/4 | 3/4 | 1/4 | 1/4 | 1/4 | 0/4 | NM | NM | NM | NM | NM |
| Neil et al., 2023 (27) | 28 | 28/28 | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM | NM |
| Kilic et al., 2019 (23); Gupta et al., 2023 (28) | 5 | 4/4 | NM | 0/4 | 2/4 | 2/4 | 0/4 | 0/4 | 2/4 | NM | NM | NM | NM | NM |
| Schallenberg et al., 2020 (32) | 19 | NM | NM | NM | NM | NM | NM | NM | NM | 0/18 | 1/18 | 2/19 | 0/19 | 2/17 |
| Total | 55 | 36/36 | Low, 4/4 | 1/8 | 5/8 | 3/8 | 1/8 | 1/8 | 2/8 | 0/18 | 1/18 | 2/19 | 0/19 | 2/17 |
| Rate | – | 100% | Low, 100% | 13% | 63% | 38% | 13% | 13% | 25% | 0% | 6% | 11% | 0% | 12% |
SMARCA4, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4; SWI/SNF, switch/sucrose nonfermenting; NM, not mentioned.
Epidemiology
Schallenberg et al. reported that 19 of 563 (3.4%) esophageal adenocarcinoma patients showed SMRACA4 protein loss on immunohistochemistry (32). However, as a newly established and rare disease entity, the exact incidence of esophageal carcinoma and SMARCA4 mutations remains unknown. Esophageal carcinoma with SMARCA4 mutations is predominant among men. Similarly, Gupta et al. reported that 80% (four out of five) of the patients (28) and Horton et al. indicated that 67% (eight out of 12) of the patients were men (31). Esophageal carcinoma with SMARCA4 mutations often affects middle-aged and older adults across a wide age range. Gupta et al. reported an age range of 48–79 years among five patients, whereas Horton et al. reported an age range of 63–79 years among 12 patients (28,31). The oldest patient with esophageal carcinoma with a SMARCA4 mutation reported in the English literature was an 85-year-old man, whereas the youngest reported patient was a 39-year-old man (29,30).
Clinical features and associated risk factors (Table 2)
Dysphagia is the most common presentation in studies reporting symptoms of esophageal carcinoma with a SMARCA4 mutation, including dysphagia, odynophagia, abdominal pain, dark stool, nausea, vomiting, and non-specific signs, including fever, fatigue, poor appetite, and weight loss (23,25-27,29). Rarely, patients initially present with symptoms of metastasis in distant organs including the lungs and liver (28).
Esophagogastroduodenoscopy (EGD) is the preferred diagnostic modality. Most esophageal carcinomas with SMARCA4 mutations occur in the distal esophagus and GEJ, and present as mass lesions on EGD (23,25-32). Horton et al. reported that of the 12 tumors, five were from the distal esophagus, two from the GEJ, two from the mid-esophagus, and three from the non-specific regions of the esophagus (31). Similarly, Gupta et al. reported that three out of five tumors were from the distal esophagus, one from the GEJ, and one from the mid to distal esophagus (28). All four patients from the series reported by Cui et al. had a tumor from the distal esophagus (25,26), whereas all four patients reported by Chang et al. had a tumor from the GEJ (30). The tumors detected on EGD varied in size, ranging from 2.5 to 10.0 cm (25,26,28-30).
Barrett’s esophagus was the most frequently associated risk factor (25,26,28,29,31). Horton et al. indicated that Barrett’s esophagus was present in 67% (eight out of 12) of patients (31), whereas both patients (100%) reported by Ahmed et al. had Barrett’s esophagus (29). Gastroesophageal reflux disease (GERD) is another commonly reported risk factor that has been reported in 20–50% of patients (25,26,28,29). Additionally, one study reported smoking in 60% (three out of five) of patients (28). Other associated risk factors included alcohol use and human immunodeficiency virus (HIV) infection in one patient (25,28,29).
Pathological and immunohistochemical features (Table 3)
Microscopically, esophageal carcinoma with SMARCA4 mutations can show different histological patterns, including epithelioid, rhabdoid, and glandular features (23,25,26,28-30) (Figures 1,2). Epithelioid features are typically present in all patients and are composed of variably cohesive neoplastic cells arranged in solid sheets, nests, cords, or trabecular formations. In high-power views, the epithelioid neoplastic cells show round to ovoid shapes, high nucleus-to-cytoplasm ratio, prominent nucleoli, atypical mitotic figures, apoptotic bodies, and tumor necrosis. Rhabdoid features are described as neoplastic cells with large eccentric nuclei, vesicular chromatin, prominent nucleoli, and cytoplasmic eosinophilic hyaline inclusions (25,28). In the study by Cui et al., three of four patients showed rhabdoid features (25), whereas four of five patients reported by Gupta et al. presented with focal rhabdoid features (28). Finally, two out of four patients reported by Cui et al. showed a rare focus of glandular differentiation (25). The histopathological results for esophageal carcinoma with SMARCA4 mutations are summarized in Table 3.
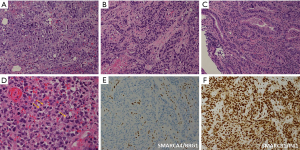
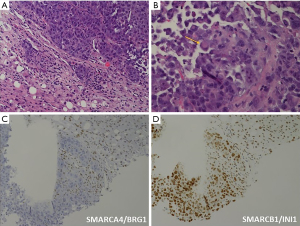
The immunohistochemical features of esophageal carcinomas with SMARCA4 mutations are summarized in Table 3 (23,25,26,28-32) (Figures 1,2). Immunohistochemistry consistently indicated SMARCA4/BRG1 protein loss in 44 out of 45 patients, except in one patient examined by Cui et al. (25). Meanwhile, SMARCB1/INI1 protein was consistently intact in all 20 patients (23,25,26,28-32). The tumor cells showed variable immunoreactivity to different cytokeratins including AE1/3 (36–100%+), Cam 5.2 (40–50%+), CK-cocktail (50%+), CK-Oscar (0–83%+), and CK7 (0–100%+), except for CK20 (23,25,26,28,30,31). The expression levels of lineage-marker varied, including CDX2 (0–33%+), GATA3 (0%+), SALL4 (50–100%+), and TTF1 (0–100%+), as well as those of squamous markers, including P40 (0–25%+) and P63 (0–100%+) (23,25,26,28,30,31). Unexpectedly, synaptophysin, a neuroendocrine marker, was detected in two out of three patients evaluated by Cui et al. and one out of two patients examined by Gupta et al.; however, other neuroendocrine markers such as chromogranin, CD56, and INSM1 were not detected (23,25,26,28). The tests for melanoma markers including SOX10 and S100 consistently showed negative results (23,25,26,28,31). The tests for lymphoid lineage markers, including CD45-LCA and CD3 for T-lymphocytes, and CD20 for B-lymphocytes also yielded negative results (23,25,26,28,31).
Molecular features and pathogenesis (Tables 3,4)
Schallenberg et al. reported that 19 of 563 (3.4%) esophageal adenocarcinoma patients showed SMRACA4 protein loss on immunohistochemistry (32). However, as a newly established and rare disease entity, the exact incidence of esophageal carcinoma and SMARCA4 mutations remains unknown. In lung cancer, the biallelic inactivation of SMARCA4 drives tumorigenesis, mainly through nonsense, frameshift, missense, and splice-site mutations; deletions; and loss of heterozygosity with a second mutation (8,33). Similarly, Neil et al. provided a detailed description of the mutation profiles of esophageal and gastric carcinomas with SMARCA4 mutations (27). Pathogenic mutations were divided into two groups. Group 1 includes nonsense, frameshift, and splice site mutations, which are predicted to lead to premature protein truncation, whereas group 2 consists of those that had been previously identified as pathogenic SMARCA4 mutational hotspots in large pan-cancer cohorts, including codons G782, G784, K785, T786, A791, P811, L815, E821, Y860, E861, E882, H884, R885, T910, P913, E920, R966, R973, R979, F1102, R1135, R1157, G1159, G1160, G1162, D1177, A1186, R1189, R1192, G1194, G1232, D1235, and R1243 (4,34,35). Unfortunately, the differences in the mutation profiles of esophageal and gastric carcinomas were not distinguished. Among the four patients evaluated by Cui et al., three harbored SMARCA4 sub-gene deep deletions, whereas one had an R1192C missense mutation. In the study by Gupta et al., the sequencing data of three patients were available, including one with copy-neutral loss of heterozygosity of SMARCA4, one with loss of chromosome 19 that harbored SMARCA4 mutations, and one with SMARCA4 subgene focal deletion.
Multiple concurrent somatic mutations were present in patients with esophageal carcinomas with SMARCA4 mutations (Tables 3,4). P53 immunohistochemistry was conducted in eighteen patients from three studies and showed mutant staining patterns ranging from 46% to 100% (25,31,32), whereas the TP53 gene mutation rates ranged from 50% to 75% (25,28,32). HER2 protein immunohistochemistry was conducted in two patients by Cui et al. and thirteen patients by Schallenberg et al., and all 15 patients exhibited negative expression of HER2 (25,32). Other somatic mutations included amplifications of CCNE1 amplification (0–25%), CDKN2A mutation (25–50%), EGFR amplification (0–25%), CTNNB1 mutation (0–25%), PTPRD mutation (0–50%), MET amplification (0%), C-myc amplification (6%), KRAS amplification (11%), GATA6 amplification (0%), and PIK3CA amplification (12%) (23,25-28,32).
In lungs, thoracic SMARCA4-deficient undifferentiated tumors were shown to be mainly immune desert tumors with no tertiary lymphoid structures, and with limited efficacy of immune checkpoint inhibitors (36). The results of immune phenotypes were limited in esophageal carcinoma with SMARCA4 mutations but showed similar trends. The tumor mutation burden was low in all four patients reported by Cui et al., while PD-L1 immunohistochemistry was performed in three patients, and one patient (33%) showed positive PD-L1 expression (combined positive score =10) (25). Mismatch repair protein immunohistochemistry was performed in seventeen patients from four studies, and all showed intact expression of MLH1, PMS2, MSH2, and MSH6 proteins (25,28,29,31).
Considerations for differential diagnosis
Owing to its rare occurrence, the pathological diagnosis of esophageal carcinoma with SMARCA4 mutations is extremely challenging, especially in small tissue fragments from biopsies, due to the following reasons. First, esophageal carcinoma with SMARCA4 mutations can show different histological patterns, including epithelioid, rhabdoid, and glandular features. Second, its immunohistochemical patterns are highly variable, especially for those widely utilized cytokeratins such as AE1/3, Cam 5.2, CK-cocktail, and CK-Oscar. Third, since it is a relatively newly established entity, SMARCA4/BRG1 immunohistochemistry is not widely available, and surgical pathologists had limited experiences of interpreting such stains. Indeed, in the first case report of esophageal carcinoma with SMARCA4 mutation conducted by Kilic et al., the initial impression was hematolymphoid malignancy owing to the weak Pax5 immunoreactivity (23). In such tumors, a meticulous assessment of the tumor morphology is of paramount importance. All reported patients exhibited epithelioid features. When these characteristics are coupled with rhabdoid features, they can serve as a significant indicator of SMARCA4-mutant carcinomas (23,25,26,28-30). Using a panel of cytokeratin immunohistochemistry assays, encompassing AE1/3, Cam 5.2, CK cocktail, CK-Oscar, and CK7, can be instrumental in confirming the epithelial/carcinoma lineage. Lymphoid markers, including CD45-LCA, CD3 for T-lymphocytes, and CD20 for B-lymphocytes, were not present in patients with esophageal carcinoma with SMARCA4 mutations (23,25,26,28,31), which helped rule out lymphoma. Finally, SMARCA4/BRG1 immunohistochemistry or SMARCA4 gene sequencing is used to obtain a definitive diagnosis.
Cui et al. described another diagnostic pitfall, in which the tumor cells of esophageal carcinoma with SMARCA4 mutations were focally positive for synaptophysin, a neuroendocrine marker (26). If synaptophysin expression is detected, the possibility of neuroendocrine carcinoma, such as small- or large-cell neuroendocrine carcinoma, should be considered. However, all other neuroendocrine markers, including chromogranin, INSM1, and CD56, were not found. The lack of typical neuroendocrine morphology, together with SMARCA4/BRG1 protein loss, helped establish a correct diagnosis. Because neuroendocrine carcinoma uses distinct chemotherapy paradigms, an extensive immunohistochemical workup should be employed to rule this out.
Other considerations for the differential diagnosis include poorly differentiated squamous cell carcinoma and melanoma. The expression profiles of squamous markers, such as P40 and P63, in esophageal carcinoma with SMARCA4 mutations vary and are usually patchy and weak; however, they are never as diffuse and strong as those observed in squamous cell carcinoma (23,25,26,28,31). Finally, melanoma markers, including SOX10 and S100, are not frequently observed in patients with esophageal carcinoma with SMARCA4 mutations (23,25,26,28,31). In Gupta et al.’s study, two patients were examined for the presence of another melanoma marker, HMB45, and both yielded a negative result (28).
In summary, awareness of the morphological and immunohistochemical profiles of esophageal carcinoma with SMARCA4 mutations is critical for making a correct diagnosis. Either SMARCA4/BRG1 protein loss in immunohistochemistry or SMARCA4 gene mutations are necessary for obtaining a definitive diagnosis.
Clinical staging, treatments, and outcome (Table 2)
Esophageal carcinoma with a SMARCA4 mutation shows universally aggressive behavior and presents at an advanced disease stage. Neil et al. showed that 7% (two out of 28) of the patients had clinical stage II, 18% (five out of 28) had clinical stage III, 18% (five out of 28) had clinical stage IVA, and 57% (sixteen out of 28) had clinical stage IVB disease (27). The staging data were also reported in other studies, including stage IB (25%), stage III (25%), stage IVA (25%), and stage IVB (25–100%) (25,28-30). Distant metastasis commonly occurs in the liver, except in one patient who had tumor metastasis in both the liver and lungs (25,28-30).
Surgical resection is the preferred treatment for esophageal carcinoma with SMARCA4 mutations. However, owing to the initial advanced disease stages, most patients are not indicated for surgery; the proportion of patients undergoing surgery is relatively low, ranging from (0–50%). The majority of patients received either chemotherapy or palliative care (25,28-30). Although the number of patients was insufficient to draw a significant conclusion, the overall patient survival outcome was still dependent on the clinical stage. One patient reported by Chang et al. initially presented with a clinical stage IB tumor, underwent surgical resection, and survived after 20 months of follow-up (30). Unfortunately, the majority of patients succumb to diseases within 1 year of initial diagnosis, with a median overall survival duration of 1–4 months (25,28-30).
Conclusions
Esophageal carcinoma with SMARCA4 mutation commonly occurs in middle-aged to older men. Barrett’s esophagus and GERD are the predominant risk factors. Dysphagia was the most common initial clinical presentation. EGD is the preferred diagnostic modality. Microscopically, the tumor cells exhibited epithelioid features mixed with variable components of rhabdoid and glandular differentiation. The tumor cells showed variable immunoreactivity for cytokeratin and sometimes weakly expressed neuroendocrine or B-cell markers (Pax5), which are potential diagnostic pitfalls. Melanoma marker tests were negative. A definitive diagnosis requires the presence of either SMARCA4/BRG1 protein loss on immunohistochemistry or SMARCA4 gene mutations. The tumor shows overly aggressive behavior and presents with advanced disease stages. Unfortunately, the majority of patients succumb to the disease within 1 year of the initial diagnosis. In conclusion, esophageal carcinoma with a SMARCA4 mutation is an overly aggressive disease, and further research on the affected molecular pathways may help improve its prognosis.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-84/rc
Peer Review File: Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-84/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-84/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Khavari PA, Peterson CL, Tamkun JW, et al. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 1993;366:170-4. [Crossref] [PubMed]
- Sarkar S, Horn G, Moulton K, et al. Cancer development, progression, and therapy: an epigenetic overview. Int J Mol Sci 2013;14:21087-113. [Crossref] [PubMed]
- Mardinian K, Adashek JJ, Botta GP, et al. SMARCA4: Implications of an Altered Chromatin-Remodeling Gene for Cancer Development and Therapy. Mol Cancer Ther 2021;20:2341-51. [Crossref] [PubMed]
- Fernando TM, Piskol R, Bainer R, et al. Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat Commun 2020;11:5551. [Crossref] [PubMed]
- Pan J, McKenzie ZM, D'Avino AR, et al. The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity-independent genomic targeting. Nat Genet 2019;51:618-26. [Crossref] [PubMed]
- Roden AC. Thoracic SMARCA4-deficient undifferentiated tumor-a case of an aggressive neoplasm-case report. Mediastinum 2021;5:39. [Crossref] [PubMed]
- Yoshida A, Kobayashi E, Kubo T, et al. Clinicopathological and molecular characterization of SMARCA4-deficient thoracic sarcomas with comparison to potentially related entities. Mod Pathol 2017;30:797-809. [Crossref] [PubMed]
- Rekhtman N, Montecalvo J, Chang JC, et al. SMARCA4-Deficient Thoracic Sarcomatoid Tumors Represent Primarily Smoking-Related Undifferentiated Carcinomas Rather Than Primary Thoracic Sarcomas. J Thorac Oncol 2020;15:231-47. [Crossref] [PubMed]
- Kolin DL, Dong F, Baltay M, et al. SMARCA4-deficient undifferentiated uterine sarcoma (malignant rhabdoid tumor of the uterus): a clinicopathologic entity distinct from undifferentiated carcinoma. Mod Pathol 2018;31:1442-56. [Crossref] [PubMed]
- Russell-Goldman E, MacConaill L, Hanna J. Primary cutaneous SMARCA4-deficient undifferentiated malignant neoplasm: first two cases with clinicopathologic and molecular comparison to eight visceral counterparts. Mod Pathol 2022;35:1821-8. [Crossref] [PubMed]
- Rooper LM, Uddin N, Gagan J, et al. Recurrent Loss of SMARCA4 in Sinonasal Teratocarcinosarcoma. Am J Surg Pathol 2020;44:1331-9. [Crossref] [PubMed]
- Agaimy A, Jain D, Uddin N, et al. SMARCA4-deficient Sinonasal Carcinoma: A Series of 10 Cases Expanding the Genetic Spectrum of SWI/SNF-driven Sinonasal Malignancies. Am J Surg Pathol 2020;44:703-10. [Crossref] [PubMed]
- Romero OA, Setien F, John S, et al. The tumour suppressor and chromatin-remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. EMBO Mol Med 2012;4:603-16. [Crossref] [PubMed]
- Bochar DA, Wang L, Beniya H, et al. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 2000;102:257-65. [Crossref] [PubMed]
- Strobeck MW, DeCristofaro MF, Banine F, et al. The BRG-1 subunit of the SWI/SNF complex regulates CD44 expression. J Biol Chem 2001;276:9273-8. [Crossref] [PubMed]
- Li R, Zhou T, Chen S, et al. Small cell carcinoma of the ovary, hypercalcemic type (SCCOHT): a challenge for clinicopathological diagnosis. Int J Clin Exp Pathol 2019;12:2166-72. [PubMed]
- Qi W, Wang R, Chen H, et al. BRG1 promotes the repair of DNA double-strand breaks by facilitating the replacement of RPA with RAD51. J Cell Sci 2015;128:317-30. [PubMed]
- Qi W, Chen H, Lu C, et al. BRG1 Promotes chromatin remodeling around DNA damage sites. Anim Cells Syst (Seoul) 2018;22:360-7. [Crossref] [PubMed]
- Hays E, Nettleton E, Carter C, et al. The SWI/SNF ATPase BRG1 stimulates DNA end resection and homologous recombination by reducing nucleosome density at DNA double strand breaks and by promoting the recruitment of the CtIP nuclease. Cell Cycle 2020;19:3096-114. [Crossref] [PubMed]
- Kurashima K, Kashiwagi H, Shimomura I, et al. SMARCA4 deficiency-associated heterochromatin induces intrinsic DNA replication stress and susceptibility to ATR inhibition in lung adenocarcinoma. NAR Cancer 2020;2:zcaa005. [Crossref] [PubMed]
- Gupta M, Concepcion CP, Fahey CG, et al. BRG1 Loss Predisposes Lung Cancers to Replicative Stress and ATR Dependency. Cancer Res 2020;80:3841-54. [Crossref] [PubMed]
- Wu Q, Lian JB, Stein JL, et al. The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics 2017;9:919-31. [Crossref] [PubMed]
- Kilic AI, Mirza K, Mehrotra S, et al. A BAFfling liver aspirate: Metastatic high grade SMARCA4 deficient undifferentiated gastroesophageal junction carcinoma masquerading as a hematolymphoid malignancy. Diagn Cytopathol 2019;47:725-32. [Crossref] [PubMed]
- WHO Classification of Tumours Editorial Board. Digestive System Tumours: WHO Classification of Tumours. 5th ed. Geneva: World Health Organization; 2019.
- Cui M, Lemmon K, Jin Z, et al. Esophageal carcinoma with SMARCA4 mutation: Unique diagnostic challenges. Pathol Res Pract 2023;248:154692. [Crossref] [PubMed]
- Cui M, Uboha N. Undifferentiated Carcinoma of Esophagus with SMARCA4 Deletion Expressing Synaptophysin: A Potential Diagnostic Pitfall. Int J Surg Pathol 2024;32:356. [Crossref] [PubMed]
- Neil AJ, Zhao L, Isidro RA, et al. SMARCA4 Mutations in Carcinomas of the Esophagus, Esophagogastric Junction, and Stomach. Mod Pathol 2023;36:100183. [Crossref] [PubMed]
- Gupta S, Noona SW, Pambuccian SE, et al. Malignant undifferentiated and rhabdoid tumors of the gastroesophageal junction and esophagus with SMARCA4 loss: a case series. Hum Pathol 2023;134:56-65. [Crossref] [PubMed]
- Ahmed OT, Nam GH, Shui Y, et al. Case Series of SMARCA4-Deficient Undifferentiated Esophageal Carcinoma. Cureus 2022;14:e30874. [Crossref] [PubMed]
- Chang B, Sheng W, Wang L, et al. SWI/SNF Complex-deficient Undifferentiated Carcinoma of the Gastrointestinal Tract: Clinicopathologic Study of 30 Cases With an Emphasis on Variable Morphology, Immune Features, and the Prognostic Significance of Different SMARCA4 and SMARCA2 Subunit Deficiencies. Am J Surg Pathol 2022;46:889-906. [Crossref] [PubMed]
- Horton RK, Ahadi M, Gill AJ, et al. SMARCA4/SMARCA2-deficient Carcinoma of the Esophagus and Gastroesophageal Junction. Am J Surg Pathol 2021;45:414-20. [Crossref] [PubMed]
- Schallenberg S, Bork J, Essakly A, et al. Loss of the SWI/SNF-ATPase subunit members SMARCF1 (ARID1A), SMARCA2 (BRM), SMARCA4 (BRG1) and SMARCB1 (INI1) in oesophageal adenocarcinoma. BMC Cancer 2020;20:12. [Crossref] [PubMed]
- Le Loarer F, Watson S, Pierron G, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet 2015;47:1200-5. [Crossref] [PubMed]
- Stanton BZ, Hodges C, Calarco JP, et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet 2017;49:282-8. [Crossref] [PubMed]
- Hodges HC, Stanton BZ, Cermakova K, et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat Struct Mol Biol 2018;25:61-72. [Crossref] [PubMed]
- Gantzer J, Davidson G, Vokshi B, et al. Immune-Desert Tumor Microenvironment in Thoracic SMARCA4-Deficient Undifferentiated Tumors with Limited Efficacy of Immune Checkpoint Inhibitors. Oncologist 2022;27:501-11. [Crossref] [PubMed]
Cite this article as: Xu J, Chi Z. Esophageal carcinoma with SMARCA4 mutation: a narrative review for this rare entity. Transl Gastroenterol Hepatol 2024;9:24.


