Genetic and epigenetic determinants of non-alcoholic fatty liver disease (NAFLD) in lean individuals: a systematic review
Highlight box
Key findings
• Our systematic review identifies genetic determinants related to lipid metabolism, inflammation, and oxidative stress pathways associated with non-alcoholic fatty liver disease (NAFLD) in lean individuals.
What is known and what is new?
• While NAFLD is common in obese individuals, this study sheds light on its occurrence in lean populations. To the best of our knowledge, this is the first systematic review on the topic.
• We highlight the role of PNPLA3 and TM6SF2 gene variants in lean NAFLD and suggest potential epigenetic contributions.
What is the implication, and what should change now?
• Clinicians should consider genetic and epigenetic factors when assessing NAFLD risk in lean patients.
• Future research should explore targeted interventions for lean individuals at risk of NAFLD, focusing on the identified genetic and epigenetic markers.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a chronic liver disease characterized by the accumulation of excessive fat in the liver in the absence of significant alcohol consumption (1). It is a prevalent liver disease worldwide, affecting approximately 25% of the general population and up to 75% of individuals with obesity and type 2 diabetes (1,2). The development of NAFLD is influenced by a complex interplay of genetic and environmental factors, including dietary habits and physical activity levels (3-5). NAFLD encompasses a spectrum of liver conditions, ranging from simple steatosis to non-alcoholic steatohepatitis (NASH), which is characterized by liver inflammation, hepatocyte injury, and fibrosis (2,6). Additionally, NAFLD is associated with an increased risk of cardiovascular disease, chronic kidney disease, and mortality, making it a significant global public health concern. Thus, it is crucial to understand the underlying pathogenesis and risk factors of NAFLD to develop effective prevention and treatment strategies.
Although NAFLD is commonly associated with obesity and metabolic syndrome, it can also occur in lean individuals (7-11). The prevalence of NAFLD in lean individuals ranges from 7% to 25% globally (12-14). Researchers have categorized lean NAFLD into two subtypes: type 1, which occurs in individuals with visceral adiposity and insulin resistance, and type 2, which arises from hepatic steatosis associated with monogenic diseases (15). Type 1 lean NAFLD accounts for 70% of cases, while type 2 represents approximately 30% (16). However, the pathogenesis of lean NAFLD remains poorly understood. Studies have shown correlations between lean NAFLD and demographics, metabolic syndrome, and genetic susceptibility (12-15,17). For example, a study involving lean Asian Indians revealed a higher prevalence of insulin resistance in Asian Indian men compared to Black, Caucasian, and Hispanic men (18). In this study, the prevalence of NAFLD in lean Indians was significantly higher than in Western individuals. Notably, the clinical definition of lean NAFLD often relies on body mass index (BMI) cut-offs or percentiles, leading to variability across studies and potentially limiting the comparability and generalizability of research efforts. Overall, the mechanisms underlying NAFLD in lean individuals may differ from those in obese individuals, potentially leading to distinct clinical outcomes (19,20).
Advancements in research have identified genetic and epigenetic modifications associated with a higher incidence of NAFLD. Several genes involved in lipid metabolism, insulin resistance, inflammation, and fibrosis have been implicated in the development and progression of NAFLD (21). While many lean individuals with NAFLD share similar clinical and genetic determinants as non-lean individuals with NAFLD, some cases may be driven by rare genetic variants (15). The cumulative impact of common genetic variants, often referred to as polymorphisms, associated with NAFLD may also contribute to the manifestation of hepatic steatosis in lean individuals despite a lower BMI (15,17,22). In recent studies, factors associated with NAFLD in lean individuals included male sex, triglyceride levels, liver transaminases, visceral adiposity, and genetic variants such as rs58542926 and rs738409 (17). The PNPLA3 gene (rs738409) emerges as a key player in the genetic landscape of NAFLD. This polymorphism exhibits three distinct genotypes—CC, CG, and GG—each associated with varying degrees of susceptibility to NAFLD development and progression (23-25). Furthermore, variants in the TM6SF2 genes have consistently shown associations with NAFLD (23-25), while genetic polymorphisms in other genes, such as ADIPOR1 and IRS1, have been linked to insulin resistance and inflammation, respectively (26-28). Additionally, epigenetic modifications, including DNA methylation and histone acetylation, have emerged as potential regulators of gene expression in NAFLD (29,30). Nonetheless, there are still significant knowledge gaps regarding how these factors contribute to the development and progression of NAFLD, particularly in lean individuals.
Emerging research highlights the complex and multifactorial nature of NAFLD development in lean individuals, involving a combination of genetic and epigenetic factors. Although studies have demonstrated that lean individuals can develop NAFLD, the underlying mechanisms and biomarkers remain unclear. Several candidate genes and epigenetic modifications have been identified in current research, potentially contributing to NAFLD development in lean individuals. However, inconsistencies in findings across studies and gaps in our understanding of the genetics and epigenetics of NAFLD in this population persist. Therefore, the aim of this study is to conduct a systematic review to identify potential genetic or epigenetic biomarkers for NAFLD in lean individuals and elucidate the underlying mechanisms of NAFLD in this specific population. The findings of this study have the potential to contribute to the development of effective diagnostic and therapeutic strategies for NAFLD in lean individuals. We present this article in accordance with the PRISMA reporting checklist (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-31/rc).
Methods
Using a set of developed keywords, multiple databases were searched on April 2nd, 2023, including: (I) PubMed; (II) MEDLINE; (III) PsycINFO; (IV) Web of Science; (V) Embase; (VI) ScienceDirect; (VII) CINAHL. The protocol related to this systematic review is registered with PROSPERO (CRD42023413809).
Search strategy and selection criteria
The authors devised a set of search keywords, which were tailored to align with this study’s research query. To optimize the search within the chosen databases, Boolean operators were employed. The resulting search terms are detailed in Table S1. Those keywords were utilized to retrieve relevant literature using the previously listed seven databases.
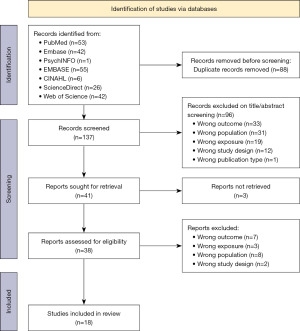
Retrieved articles (n=225) were entered into Endnote reference manager software, after which all duplicates were removed (n=88). As shown in the PRISMA flow diagram in Figure 1, two investigators (Y.A.A.A. and B.N.) independently assessed the title and abstracts of all unique search results (n=137), to determine their relevance. After eliminating articles that were deemed irrelevant at this stage (n=96), the full-text versions were obtained and assessed for eligibility. Studies were included if they: (I) were conducted and published in English language; (II) had their primary outcome reports genetic or epigenetic effects on NAFLD; (III) included lean individuals as a population; (IV) had their full-text paper available; (V) were peer-reviewed prior to final publication. Similarly, studies were excluded if they: (I) were not peer-reviewed; (II) had their full-text paper unavailable; (III) were written in a language that is not English; (IV) were designed as a review of the literature (e.g., scoping, systematic, narrative, or any other review). Table S2 outlines the full eligibility criteria for studies upon screening of the literature search results. Furthermore, in cases where a single study or dataset was documented in multiple publications, we opted for the publication that conveyed outcomes and additional pertinent information that aligned with our research query. Alternatively, we considered the publication that was published in a more appropriate format, such as an original research article as opposed to a research letter.
Data extraction
A tabulated form was prepared for data extraction after the screening stage. Data extraction was performed by one reviewer using a pre-designed table reviewed by co-authors. The data reflected the genetic and epigenetic determinants of NAFLD among lean individuals. Different dimensions were collected for each study, including: the name of the first author and the year of publication; the study design; population sociodemographic; number of lean individuals; definition of lean individuals; outcome investigated; summary of main findings; summary of main genetic/epigenetic findings.
Data presentation
The collected data, which encompasses the genetic and epigenetic factors of NAFLD in lean individuals, are arranged in a tabular format with no distinct categorization. Furthermore, the genetic and epigenetic determinants of NAFLD in lean individuals are outlined in a textual format. In addition to identifying the primary genetic and epigenetic determinants of NAFLD documented in existing literature, this presentation of data attempts to offer an overview of the studied population and exposes any gaps in the current body of knowledge.
Quality appraisal
The Newcastle Ottawa Scale (NOS) was employed to appraise the quality of the included studies. Two investigators conducted independent assessments of the risk of bias, and in instances of disagreement, a third investigator was consulted for resolution. A total of 18 non-randomized studies were evaluated for quality using the NOS tool. Other study designs were not subject to quality assessment but were included in the synthesis of data.
The NOS was developed as an assessment tool for evaluating the quality of nonrandomized studies, with its design, content, and usability aimed at incorporating quality assessments into the interpretation of meta-analytic outcomes (1). The NOS evaluation system comprises of seven categories for scoring and adopts a star system that is subcategorized into three main themes: selection (maximum of four stars across four sub-categories), comparability (maximum of two stars across one sub-category), and outcome (maximum of three stars across two sub-categories). The sub-categories cover aspects such as sample representativeness, sample size, comparability between respondents and non-respondents, ascertainment of exposure, comparability with respect to participant distribution and analyses, assessment of outcome, and statistical tests. A full list of adapted questions from the NOS is shown in Appendix 1.
Ethical approval
In the context of this systematic review, ethical approval is deemed unnecessary as the data utilized was procured from pre-existing literature. The studies encompassed within this review were subjected to ethical review and clearance by their respective primary investigators prior to data collection.
Results
Search results
The systematic literature search yielded a total of 225 articles, obtained from the following databases as follows: 53 articles from PubMed, 42 articles from MEDLINE, 1 article from PsycINFO, 55 articles from Embase, 6 articles from CINAHL, 26 articles from Science Direct, and 42 articles from Web of Science. After removing duplicate studies (n=88), a total of 137 articles were screened based on title and abstract. At this stage, a total of 96 studies were excluded. Reasons for exclusion at this stage included studies investigating wrong outcome (n=33), wrong population (n=31), wrong exposure (n=19), wrong study design (n=12), and wrong publication type (n=1). Full-text versions of 41 articles were sought, and 38 full-text studies were reviewed (three studies were excluded given unavailability of full-text versions). Reasons for exclusion at this stage included wrong outcome (n=7), wrong exposure (n=3), wrong population (n=8), wrong study design (n=2). Consequently, a total of 18 studies were included in the qualitative data synthesis for this systematic review.
Study characteristics
A total of 18 studies were included in this review. The study sample sizes ranged from 53 participants (2) to 5,338 participants (3), with Li et al. [2023] having the highest number of lean NAFLD patients included in their analysis (n=2,012). Studies were conducted globally, including Japan (4,5), Sri Lanka (6), Australia (7), India (8), Italy (9), and Sweden (10). Furthermore, the majority of included studies were nonrandomized, including cross-sectional study design (n=12) and cohort study design (n=6).
Quality appraisal
The outcomes of the assessment of study quality are shown in Tables S3,S4. For all nonrandomized studies (n=18), the NOS protocol detailed in the methods section was used for the evaluation. Overall, the majority of studies had a moderate quality, with an average of approximately 7 stars (range, 5–9 stars). There were two studies with 5 stars, three studies with 6 stars, six studies with 7 stars, one study with 8 stars, four studies with 9 stars, and two studies with 10 stars.
Genetic and epigenetic determinants
The systematic review of studies investigating the genetic and epigenetic determinants of NAFLD in lean individuals is summarized in Table 1. The review revealed a range of conclusions across the selected studies, with a predominant focus on reporting genetic determinants. However, despite the heterogeneity in study design and sample characteristics, several common genetic and epigenetic associations emerged across multiple populations.
Table 1
| Author, year | Study design | Population sociodemographic | No. of lean individuals | % of lean individuals with mutation/gene | Definition of lean individuals | Outcomes investigated | Main findings | Main genetic/epigenetic findings |
|---|---|---|---|---|---|---|---|---|
| Buzova et al., 2020 (2) | Prospective cohort study | The study included a cohort of 53 patients diagnosed with MAFLD, comprising 38 who were classified as overweight, 15 categorized as lean, alongside 120 samples collected from control participants | N=15 | N/A | BMI <25 kg/m2 | Circulating nucleosome levels in the blood, as a marker of epigenetics of disease | Circulating nucleosome levels associated poorly with MAFLD in the absence of obesity | A significant depletion of the levels of histone variants macroH2A1.1 and macroH2A1.2 in the serum of lean MAFLD patients, either individually or in complex with H2B |
| Adams et al., 2012 (19) | Candidate gene approach | A total of 951 17-year-old individuals underwent hepatic ultrasonography, as well as comprehensive assessments of anthropometric measurements and biochemical profiles, followed by DNA extraction and genotyping involving 57 SNPs within seven genes associated with lipid metabolism, namely ApoB100, ATGL, ABHD5, MTTP, CETP, SREBP-1c, and PPARα | N/A | The prevalence of this condition was observed to be approximately 10–15% among individuals classified as lean heterozygotes, while it was notably lower, around 3–5%, among those classified as lean wild-types | N/A | “This study aimed to use a candidate gene approach to examine the association between NAFLD and single nucleotide polymorphisms (SNPs) in lipid metabolism genes in the adolescent population-based Western Australian Pregnancy (Raine) Cohort.” | Cholesteryl ester transfer protein gene polymorphisms are associated with an elevated risk of FLD in adolescent females. This risk is independent of adiposity in homozygotes, highlighting that even lean individuals face a significant risk of FLD | In females, two SNPs in linkage disequilibrium from the “CETP gene were associated with NAFLD: rs12447924 (OR 2.16, 95% CI: 1.42–3.32, P=0.0003) and rs12597002 (OR 2.22, 95% CI: 1.46–3.41, P=0.0002). In lean homozygotes, the probability of NAFLD was over 30%, compared with 10–15% in lean heterozygotes and 3–5% in lean wild types.” |
| Wei et al., 2015 (11) | Cross-sectional study | A total of 911 community subjects were randomly recruited from the census database of the Hong Kong Government. Intrahepatic triglycerides and liver fibrosis were assessed by proton-magnetic resonance spectroscopy and transient elastography, respectively | N/A | 78.4% | The Asian BMI cutoff of 25 kg/m2 was used to define non-obese NAFLD | “The study investigated the epidemiology and severity of non-obese NAFLD. Clinical assessment included gene polymorphism measurements.” | When compared to obese NAFLD patients, non-obese NAFLD patients exhibited comparable intrahepatic triglyceride content (median 9.8% vs. 9.9%; P=0.100). However, they displayed lower levels of cytokeratin-18 fragments (149 vs. 182 IU/L; P=0.019) and reduced liver stiffness (4.6 vs. 5.6 kPa; P<0.001) | “The G allele at the patatin-like phospholipase domain-containing protein 3 gene (PNPLA3 rs738409) was more common in non-obese than obese NAFLD patients (78.4% vs. 59.8%; P=0.001). Obesity, high hemoglobin A1c, insulin resistance, hyperferritinemia, and the PNPLA3 G allele were independent factors associated with NAFLD in non-obese subjects.” |
| Shen et al., 2014 (12) | Cross-sectional study | “Liver fat and fibrosis were assessed by proton-magnetic resonance spectroscopy and transient elastography in 920 subjects from a population screening project (251 had NAFLD). Mean BMI for the sample was 22.8.” | N/A | N/A | N/A | To examine the association among PNPLA3 gene polymorphism, dietary pattern, metabolic factors and NAFLD | The presence of the G allele emerged as a predictor of NAFLD, irrespective of nutrient intake and other metabolic factors, with adjusted ORs of 2.00 for CG and 2.68 for GG when compared to CC genotype individuals. Notably, in individuals without metabolic syndrome, the G allele exhibited an even stronger association with NAFLD diagnosis, with adjusted ORs of 2.22 for CG and 3.39 for GG in comparison to CC genotype individuals | The G allele in PNPLA3 rs738409 increases the risk of NAFLD in the general population, especially in subjects without metabolic syndrome, independent of dietary pattern and metabolic factors |
| Petersen et al., 2010 (8) | Validation study | 95 healthy Asian Indian men | N/A | 30% | N/A | The aim of this investigation was to assess whether the C-482T and T-455C polymorphisms in the APOC3 gene are linked to NAFLD and insulin resistance in lean individuals | “Individuals with nonalcoholic fatty liver disease exhibited significant insulin resistance. This relationship was further validated in a study involving non-Asian Indian men, which affirmed the association between APOC3 variant alleles and nonalcoholic fatty liver disease.” | Carriers of APOC3 variant alleles (C-482T, T-455C, or both) exhibited a 30% rise in fasting plasma apolipoprotein C3 levels compared to wild-type homozygotes. Moreover, they experienced a 60% increase in fasting plasma triglyceride concentrations, a twofold elevation in plasma triglyceride and retinyl fatty acid ester concentrations following an oral fat-tolerance test, and a 46% reduction in plasma triglyceride clearance. Notably, the prevalence of NAFLD was 38% among individuals carrying variant alleles, while it remained at 0% among wild-type homozygotes (P<0.001) |
| Niriella et al., 2019 (6) | Community-based follow-up study and case-control study | 2,985 participants | 120 (4.0%) had lean NAFLD | N/A | Lean (BMI <23 kg/m2) | This study investigated and compared the clinical characteristics, body composition, metabolic associations and outcomes, and other risk factors among individuals with lean (BMI <23 kg/m2) NAFLD, non-lean (BMI ≥23 kg/m2) NAFLD and those without NAFLD | While lean NAFLD represents a minority within the spectrum of NAFLD cases, the risk of developing new metabolic comorbidities in these individuals is comparable to that observed in non-lean NAFLD cases | “A PNPLA3 variant showed association with lean NAFLD in the studied population. Therefore, lean NAFLD also warrants careful evaluation and follow-up.” |
| Lin et al., 2022 (14) | Cross-sectional study | 904 community subjects | 529 (58.5%) | 30.3% | Lean (<23 kg/m2) | The researchers aimed to investigate the relationship between genetic risk variants and the susceptibility as well as the severity of NAFLD in individuals across different categories of body weight, including lean, overweight, and obese individuals | The PNPLA3 rs738409 gene polymorphism exerts a more pronounced impact on liver fat levels in Asian individuals who are lean compared to those who are overweight or obese | Among individuals with NAFLD, it was observed that lean subjects (30.3%) were more likely to carry the PNPLA3 rs738409 GG genotype in comparison to overweight (17.9%) and obese subjects (17.4%) (P=0.003). When compared to the CC genotype, the GG genotype was associated with the highest increase in the risk of NAFLD among lean subjects (OR 6.04), as opposed to overweight (OR 3.43, 95% CI: 1.06, 11.14) and obese subjects (OR 2.51, 95% CI: 0.93, 6.78). Furthermore, it was noted that the TM6SF2 rs58542926 TT genotype was linked to reduced serum triglyceride levels solely in lean subjects. However, there was no evidence of a gene-BMI interaction effect for the other gene polymorphisms studied |
| Honda et al., 2016 (5) | Cross-sectional study | 540 NAFLD patients (134 non-obese and 406 obese) and 1,012 control subjects (782 non-obese and 230 obese) | N/A | N/A | N/A | Investigating genetic and other clinical parameters in non-obese and obese NAFLD | This study provided evidence that the risk factors contributing to the onset and advancement of NAFLD varied between non-obese and obese patients. Specifically, they found a strong association between the PNPLA3 rs738409 gene variant and the development as well as progression of NAFLD specifically in non-obese individuals | “Non-obese NAFLD subjects had a higher rs738409 GG genotype than obese NAFLD. Multiple logistic regression analysis indicated that the odds ratios of T2DM and rs738409 GG genotype for NAFLD were higher in non-obese than in obese groups. In non-obese NAFLD, rs738409 GG genotype was associated with lobular inflammation, hepatocyte ballooning and NAFLD activity score. In obese NAFLD, BMI and T2DM but not rs738409 GG genotype were associated with severity of histology.” |
| Hagström et al., 2017 (10) | Cohort study | 646 patients with biopsy‐proven NAFLD | “Lean NAFLD was seen in 19% of patients” | N/A | lean (BMI <25.0 kg/m2) | “The overall aim of the study was to investigate the long‐term prognosis in a large cohort of patients with biopsy proven lean NAFLD with a prolonged follow‐up time.” | During a mean follow-up duration of 19.9 years (ranging from 0.4 to 40 years), involving a cumulative period of 12,631 person-years, individuals with lean NAFLD did not demonstrate an increased risk of overall mortality in comparison to overweight patients (with a hazard ratio of 1.06 and a non-significant P value of 0.73). Nevertheless, it was noted that lean NAFLD patients had a significantly higher risk of developing severe liver disease, as evidenced by a hazard ratio of 2.69 and a statistically significant P value of 0.007, when compared to their overweight counterparts | “In a cohort of non-obese patients with biopsy-proven NAFLD, it was observed that PNPLA3 and TM6SF2 polymorphisms did not show any significant associations with the histological severity of NAFLD or disease progression.” |
| Fracanzani et al., 2017 (9) | Cohort study | 669 consecutive patients with biopsy proven NAFLD | n=143 | 4% | Lean NAFLD was defined as having a BMI less than 25 kg/m2 | The primary objective of this study was to investigate differences between lean NAFLD and NAFLD in overweight and obese individuals, identify factors associated with the severity of liver and cardiovascular disease, and evaluate the effects of visceral obesity | A significantly lower proportion of individuals diagnosed with lean NAFLD (143 patients; 43 women; mean age, 46±13 years) displayed hypertension (P=0.001), diabetes (P=0.0001), and metabolic syndrome (P=0.0001) in comparison to their counterparts with NAFLD who were overweight or obese (526 patients; 149 women; mean age, 49±12 years). Furthermore, individuals with lean NAFLD exhibited markedly lower prevalence rates of NASH (17% vs. 40% in obese or overweight NAFLD patients; P=0.0001), fibrosis at stage F2 or higher (17% vs. 42%; P=0.0001), and carotid plaques (27% vs. 39%; P=0.03). Additionally, those with lean NAFLD had a thinner carotid intima-media measurement (0.74±0.1 mm) compared to their obese or overweight NAFLD counterparts (0.84±0.3 mm; P=0.0001) | “There was no significant difference in the proportions of patients with rs738409 C>G in PNPLA3, but a significantly greater proportion of patients with lean NAFLD carried rs58542926 C>T in TM6SF2 (4%) than obese or overweight individuals with NAFLD (0.3%; P=0.001).” |
| Feldman et al., 2017 (7) | Cross-sectional study | 187 subjects | N=71 | N/A | Lean healthy (BMI ≤25 kg/m2) | The study aimed to investigate clinical, genetic, metabolic and lifestyle characteristics in lean Caucasian subjects with NAFLD | Lean NAFLD subjects had fasting insulin concentrations similar to lean healthy subjects but had markedly impaired glucose tolerance | Lean NAFLD subjects had a higher rate of the mutant PNPLA3 CG/GG variant compared to lean controls (P=0.007) |
| Zeng et al., 2020 (18) | Cross-sectional study | The study encompassed a total of 2,715 subjects who had undergone routine health examinations | 810 lean participants with a normal WC | N/A | “A BMI less than 23 kg/m2 and a WC less than 80 cm in females or 90 cm in males were used to define the lean population with a normal WC.” | The study aimed to investigate the prevalence, clinical characteristics, risk factors, and potential indicators for NAFLD in lean Chinese adults with a normal WC | Among the 810 lean participants with a normal WC, 142 individuals (17.5%) were diagnosed with NAFLD based on established criteria. It's noteworthy that factors including waist-height ratio, hemoglobin levels, platelet counts, and triglyceride levels demonstrated significant associations with the presence of NAFLD in this particular cohort. Furthermore, the researchers identified an optimal cut-off value for the FLI score when screening for NAFLD among lean subjects with a normal WC, which was determined to be 25.15. This cut-off yielded a sensitivity of 77.8% and a specificity of 75.9% | “There was no significant difference in the single-nucleotide polymorphisms in the SIRT1, APOC3, PNPLA3, AGTR1, and PPARGC1A genes between lean subjects with and without NAFLD (P<0.05).” |
| Yoshida et al., 2020 (4) | Cross-sectional study | In stage I, 275 metabolically healthy normal‐weight patients with NAFLD were compared with 1,411 non‐NAFLD. In stage II, 9,726 members of the general population | N=275 | N/A | Non-obese NAFLD (BMI <25 kg/m2) | The aim of the study was to identify NAFLD‐associated loci in Japanese patients | HLA was identified as a previously unrecognized genetic locus linked to susceptibility to NAFLD, and it is possible that this association is influenced by changes in gut microbiota | “A minor allele of the secondary lead SNP in chr6, rs2076529, was significantly associated (odds ratio [OR], 1.19; 95% confidence interval [CI], 1.11–1.28; P=2.10E−06) and the lead SNP in chr7 was weakly associated (OR 1.15; 95% CI, 1.04–1.27; P=6.19E−03) with increased NAFLD risk. Imputation‐based typing of HLA showed a significant difference in the distribution of HLA‐B, HLA‐DR‐beta chain 1 (DRB1), and HLA‐DQ‐beta chain 1 (DQB1) alleles in lean NAFLD GWAS. Next‐generation sequence‐based typing of HLA in 5,649 members of the general population replicated the significant difference of HLA‐B allele distribution and the significant increase of the HLA‐B*54:01 allele in whole NAFLD. Fecal metagenomic analysis of 3,420 members of the general population showed significant dissimilarity in beta‐diversity analysis of rs2076529 and HLA‐B*54:01 allele carriers from noncarriers. Veillonellaceae was increased but Verrucomicrobia was decreased in rs2076529 minor allele and HLA‐B*54:01 allele carriers as in NAFLD.” |
| Stasinou et al., 2022 (13) | Cross-sectional, case-control study | The study involved the monitoring of 182 children and adolescents within the Pediatric Gastroenterology Unit of the Fourth Department of Pediatrics at the Aristotle University of Thessaloniki | “14 were boys and 13 were girls (mean age 9.15±3.86 years).” | N/A | BMI <85th percentile | “The study aimed to determine the association of PNPLA3 variants with NAFLD susceptibility in obese and nonobese Greek children and adolescents.” | “A significant correlation was shown between the rs738409 polymorphism (CG and GG genotypes) and the rs2896019 polymorphism (TG genotype) with the development of hepatic steatosis (P<0.001).” | The incidences of rs738409 GG, rs738409 CG and rs2896019 TG genotypes were found to be increased in patients with hepatic steatosis (obese and nonobese), but not in obese patients without liver disease. The combined expression of the 2 polymorphisms was associated with a lower age of diagnosis of hepatic steatosis in nonobese patients |
| Stanislawski et al., 2020 (17) | Prospective cohort study | 347 children aged 12.5–19.5 years | N/A | Genetic risk factors were more important among lean individuals (2.7% for cardio metabolic markers vs. 12.6% for rs738409 and 4.4% for GRS | BMI-for-age: underweight (defined as <5th percentile) | The aim of this study was to evaluate the significance of both genetic and non-genetic risk factors in relation to hepatic fat content among a diverse population of young individuals | PNPLA3 rs738409 and the GRS were both linked to HFF in Hispanic youth (β=0.39; 95% CI: 0.16, 0.62; P value=0.001; and β=0.20; 95% CI: 0.05, 0.34; P=0.007, respectively). However, these associations were not observed in non-Hispanic White youth (β=0.04; 95% CI: −0.18, 0.26; P =0.696; and β=0.03; 95% CI: −0.09, 0.14; P=0.651, respectively) | “Cardiometabolic risk factors explained more of the variation in HFF than genetic risk factors among non-lean Hispanic individuals (27.2% for cardiometabolic markers versus 6.4% for rs738409 and 4.3% for the GRS), and genetic risk factors were more important among lean individuals (2.7% for cardiometabolic markers versus 12.6% for rs738409 and 4.4% for GRS).” |
| Li et al., 2023 (3) | Post-doc analysis of a cross-sectional population study | 5,338 community subjects | n=2,012 | N/A | Lean is defined as having BMI <23 kg/m2 | The study investigated the prevalence and clinical metabolic characteristics of lean individuals with NAFLD among elderly Chinese population and assessed the relevance of lipid markers and genetic variation | The study’s findings indicated that the frequency of the C allele for rs2279026, the G allele for rs2279028, the C allele for rs780093, and the C allele for rs1260326 were notably higher in individuals with obese NAFLD compared to those with lean NAFLD (P<0.05) | The study noted a correlation between the CC genotype of rs1421085, the TT genotype of rs3751812, the AA genotype of rs8050136, and the AA genotype of rs9939609 within the FTO gene and low-density lipoprotein levels (P<0.05) |
| Chatterjee et al., 2021 (16) | Exome-wide approach | 244 participants | N/A | N/A | N/A | The study used an exome-wide approach to identify genetic determinants of HFC in India | The study identified 4 significantly associated SNPs (rs738409 and rs2281135 (PNPLA3), rs3761472 (SAMM50), rs17513722 (FAM161A) and rs4788084), with HFC after adjusting for the effects of covariates (P<0.0005). rs738409, rs2281135 (PNPLA3), and rs3761472 (SAMM50) were associated with hepatocyte ballooning, lobular and portal inflammation and NASH (P<0.05) | In every subset of participants in this study—lean, overweight and obese—mean HFC was significantly higher among individuals who possessed the risk alleles for rs738409 (G) and rs4788084 (A) SNPs, compared with those who did not |
| Chahal et al., 2022 (15) | Cross-sectional study | 1,007 patients with NAFLD | N=136 with BMI <25 kg/m2 | N/A | BMI <25 kg/m2 | The study utilized the UK Biobank to carry out a cross-sectional investigation aimed at identifying distinguishing characteristics between lean NAFLD and NAFLD in individuals who are overweight or obese | In patients with a BMI less than 25, factors associated with NAFLD encompassed male sex, white blood cell count, red blood cell count, triglyceride levels, ALT levels, creatinine levels, visceral adipose tissue, as well as the genetic variants rs58542926 T and rs738409 G. Conversely, in patients with a BMI greater than or equal to 25, factors linked to NAFLD included male sex, waist circumference, HDL cholesterol levels, triglyceride levels, serum glucose levels, ALT levels, creatinine levels, urate levels, visceral adipose tissue, and the genetic variants rs1260326 T, rs1044498 C, rs58542926 T, and rs738409 G | “For lean patients, the study generated prediction score that had an AUC of 0.92, sensitivity of 0.90 and specificity of 0.81. For overweight or obese patients, the prediction score had an AUC of 0.86, sensitivity of 0.87 and specificity of 0.70.” |
MAFLD, metabolic-associated fatty liver disease; BMI, body mass index; SNP, single nucleotide polymorphism; NAFLD, non-alcoholic fatty liver disease; OR, odds ratio; CI, confidence interval; T2DM, type 2 diabetes; NASH, non-alcoholic steatohepatitis; WC, waist circumference; GRS, genetic risk score; HFF, hepatic fat fraction; HFC, hepatic fat content; ALT, alanine transaminase.
Limited evidence of epigenetic determinants in lean NAFLD: insights from a prospective cohort study
Firstly, one study investigated epigenetic determinants of NAFLD in lean individuals. Buzova et al. [2020] conducted a prospective cohort design conducted in Rome, Italy, and included a total of 53 NAFLD patients, including 15 lean patients with a BMI <25 kg/m2 (2). The main outcome investigated was circulating nucleosome levels in the blood, as a marker of epigenetic modifications in the disease. The results showed that circulating nucleosome levels were poorly associated with NAFLD in the absence of obesity. Furthermore, a significant depletion of the levels of histone variants macroH2A1.1 and macroH2A1.2 was observed in the serum of lean NAFLD patients, either individually or in complex with H2B. These findings suggest that histone modifications may play a role in the pathogenesis of NAFLD in lean individuals, providing potential targets for future therapeutic interventions. It is noteworthy that Buzova et al.’s study is the only one among the selected studies that directly investigated epigenetic determinants of NAFLD in lean individuals. This highlights the limited literature on this topic, particularly in comparison to studies investigating genetic determinants. The identified epigenetic modifications, particularly the depletion of histone variants, warrant further investigation to fully understand their role in the pathogenesis of NAFLD in lean individuals.
PNPLA3 genetic variant and NAFLD risk in lean individuals: findings from diverse populations
With limited studies investigating epigenetic determinants in NAFLD among lean individuals, most of the selected studies focused on reporting genetic determinants associated with the development of the disease among lean individuals. Numerous studies have investigated the relationship between PNPLA3 polymorphism and NAFLD in lean patients. One such study by Wei et al. (11) utilized proton-magnetic resonance spectroscopy and transient elastography techniques to examine 911 community subjects from Hong Kong with a non-obese BMI cutoff of 25 kg/m2. Results from this study revealed that the G allele at the PNPLA3 rs738409 gene was more common in non-obese NAFLD patients than in obese NAFLD patients (78.4% vs. 59.8%; P=0.001), indicating a genetic determinant of the disease. Similarly, and among a cohort of 920 lean individuals, Shen et al. (12) demonstrated an association between PNPLA3 gene polymorphism, dietary pattern, metabolic factors, and NAFLD. The study found that the presence of the G allele was an independent predictor of NAFLD (adjusted odds ratio to CC: CG, 2.00; GG, 2.68), even after controlling for nutrient intake and other metabolic factors. Additionally, among individuals who did not have metabolic syndrome, the presence of the G allele exhibited a notably stronger association with NAFLD, with adjusted odds ratios of 2.22 for CG and 3.39 for GG compared to CC genotype individuals. These combined results indicate that the G allele of PNPLA3 rs738409 elevates the susceptibility to NAFLD in the overall population, especially in lean individuals lacking metabolic syndrome, regardless of their dietary patterns and metabolic profile. The conclusion that rs738409 genotype may be a risk factor allele among lean NAFLD patients was also supported by other studies conducted among different populations, including lean NAFLD Greek (13), Sri Lankan (6) and Japanese (5) populations. Among the Greek cohort of children and adolescents, an important finding emerged, indicating a notable association between the rs738409 polymorphisms (CG and GG genotypes) and the rs2896019 polymorphism (TG genotype) with the onset of hepatic steatosis (13). Additionally, and for the first time, this study investigates the connection between the rs2896019 polymorphism and hepatic steatosis, encompassing both obese and nonobese pediatric patients (13). Finally, in the Japanese cohort of lean NAFLD patients investigated by Honda et al., The presence of the rs738409 GG genotype was linked to findings of lobular inflammation, hepatocyte ballooning, and an elevated NAFLD activity score (5).
Moreover, Feldman et al. investigated the prevalence of PNPLA3 risk alleles in a cohort of 187 Austrian participants and noted a heightened occurrence of these alleles in lean individuals with NAFLD when compared to the lean control group, and the frequency was similar to that observed in NAFLD patients with obesity (7). In a retrospective cohort study conducted in Italy, Fracanzani et al. examined 669 consecutive patients with biopsy-confirmed NAFLD across three liver centers. Despite no substantial differences in the frequency of the rs738409 C>G polymorphism in PNPLA3 among patients, the presence of the rs738409 (PNPLA3) risk allele was found to be an independent factor associated with the occurrence of NASH and a fibrosis score of ≥2 in individuals with a lean body composition (9). Differently from Wei et al., in this study, no disparities were identified in the occurrence of the PNPLA3 polymorphism (both heterozygous and homozygous) between lean individuals with NAFLD and those with overweight/obese NAFLD. However, it’s worth noting that individuals carrying this polymorphism in this cohort faced an increased risk of experiencing more pronounced liver damage. Furthermore, this study also found a significant association between the presence of the rs58542926 risk allele and NAFLD in lean individuals, which was not reported previously. Among their sample, approximately 4% of lean NAFLD patients carried the rs58542926 allele compared to 0.3% of non-lean NAFLD patients.
BMI-dependent effects of TM6SF2 genetic variants on NAFLD susceptibility
Involving 904 Japanese participants residing in the community, the prevalence of NAFLD was found to be 12.4%, with proportions of 41.4% in the lean group, 12.4% in the overweight group, and 59.1% in the obese group. Lin et al. (14) reported that among those with NAFLD, exhibited a twofold higher likelihood of harboring the PNPLA3 rs738409 GG genotype when compared to their overweight and obese counterparts, a trend that aligns with consistent observations reported in previous studies. Compared with the CC genotype, the GG genotype was associated with the greatest increase in the risk of NAFLD in lean subjects, compared with overweight, and obese subjects. Upon stratification by BMI, no disparities in risk were observed concerning the MBOAT7 or TM6SF2 NAFLD risk alleles. Interestingly, this study also reiterates Fracanzani et al.’s (9) findings regarding rs58542926 polymorphism, as they found the study revealed that the TM6SF2 rs58542926 TT genotype was linked to lower serum triglyceride levels exclusively in lean individuals. There was no observed gene-BMI interaction effect for the other gene polymorphisms. In a separate investigation, Chahal et al. employed the UK Biobank for a cross-sectional study, aiming to identify characteristics that differentiate lean NAFLD from NAFLD in individuals who are overweight or obese (15). Through this cross-sectional study, many factors were identified to be associated with lean NAFLD, including rs58542926 T, and rs738409 G genotypes. Furthermore, and among 146 Indian patients with liver-biopsy diagnosed NAFLD, employing an exome-wide methodology to pinpoint genetic factors influencing hepatic fat content (HFC), the study disclosed that individuals carrying the risk alleles for the rs738409 (G) and rs4788084 (A) single nucleotide polymorphisms (SNPs) had notably elevated mean HFC levels compared to those lacking these alleles. (16). This included a stratified analysis among lean individuals. In a multiethnic cohort of 347 children aged 12.5–19.5 years, Stanislawski et al. (17) investigated the relationship between genetic factors and hepatic fat fraction (HFF). Variations in the association based on ethnicity (Hispanic vs. non-Hispanic white) and BMID category were also compared. Among Hispanics, it was shown that the PNPLA3 rs738409 was associated with HFF, but not among non-Hispanic white youth. Furthermore, cardiometabolic risk factors explained more of the variation in HFF than genetic risk factors among non-lean Hispanic individuals. However, among lean individuals, genetic factors were more important than cardiometabolic markers (17).
Contrasting findings in lean individuals with NAFLD: genetic and clinical characteristics
Two studies, Zeng et al. (18) and Hagström et al. (10), reported findings that do not support the presence of relevant associations in relation to the genetic and clinical characteristics of lean individuals with NAFLD.
Zeng et al. [2020] conducted a cross-sectional study involving 2,715 subjects who underwent routine health examinations, with a focus on investigating the prevalence, clinical characteristics, risk factors, and potential indicators for NAFLD among lean Chinese adults with a normal waist circumference (WC). Out of the 810 lean participants with a normal WC, 142 individuals (17.5%) fulfilled the criteria for NAFLD. Key factors such as waist-height ratio, hemoglobin levels, platelet counts, and triglyceride levels were identified as significant factors associated with NAFLD presence in this group. However, there were no significant differences detected in the single-nucleotide polymorphisms of the SIRT1, APOC3, PNPLA3, AGTR1, and PPARGC1A genes between lean subjects with and without NAFLD in the study. Similarly, in a cohort study by Hagström et al. [2017], 646 patients with biopsy proven NAFLD were followed up over a mean duration of 19.9 years. The study aimed to investigate the long-term prognosis of patients with lean NAFLD. Surprisingly, lean NAFLD was observed in 19% of the patients, and the study found no increased risk for overall mortality in these patients compared to overweight individuals. However, patients with lean NAFLD did exhibit an increased risk for the development of severe liver disease. Additionally, the study did not find any associations between PNPLA3 and TM6SF2 polymorphisms and the histological severity of NAFLD or disease progression in this cohort of non-obese patients with biopsy proven NAFLD.
Associations of other alleles: CETP and APOC3 genetic variants in NAFLD risk among lean individuals
Furthermore, other genetic risk factors were reported in the literature, despite less frequency. For instance, the cholesteryl ester transfer protein gene was investigated among NAFLD patients in Australia. It was speculated that polymorphism in the gene may increase the risk of fatty liver. Using a candidate gene approach, Adams et al., found that two SNPs in linkage disequilibrium from the CETP gene were associated with FLD among females, including rs12447924 and rs12597002. Interestingly, this association was independent of adiposity in homozygous patients. In turn, it is plausible to claim that lean individuals who are homozygotes for the cholesteryl ester transfer protein gene polymorphism are placed at a significant risk of NAFLD (19). Moreover, another study revealed that Indian men with NAFLD were found to carry two gain-of-function SNPs within the gene encoding APOC3. Among a sample of 95 Asian Indian men, Carriers of the APOC3 variant alleles (C-482T, T-455C, or both) had a 30% increase in the fasting plasma apolipoprotein C3 concentration, as compared with the wild-type homozygotes. Additionally, the prevalence of nonalcoholic fatty liver disease was 38% among variant-allele carriers and 0% among wild-type homozygotes (P<0.001). The subjects with nonalcoholic fatty liver disease had marked insulin resistance (8).
The role of HLA locus and gut microbiota
Yoshida et al. (4) conducted a cross-sectional study to investigate the genetic determinants of NAFLD among lean individuals. The study conducted a two-stage analysis in Japanese patients, using a genome-wide association study (GWAS) to compare 275 metabolically healthy normal-weight patients with NAFLD to 1,411 non-NAFLD controls in stage I. In stage II, the study assessed human leukocyte antigen (HLA) in chromosome 6, microRNA (MIR) MIR548F3 in chr7, myosin light chain 2 (MYL2) in chr12, and glycoprotein precursor (GPC)6 in chr13, as suggested by the GWAS, by SNP association analysis of whole NAFLD against non-NAFLD in 9,726 members of the general population. Their findings revealed that the HLA locus, specifically the rs2076529 SNP in chromosome 6, was significantly associated with an increased risk of NAFLD. Furthermore, HLA typing and next-generation sequencing further supported the significance of HLA-B alleles, particularly the HLA-B54:01 allele, in lean NAFLD patients. Fecal metagenomic analysis indicated differences in the gut microbiota composition of individuals carrying the rs2076529 minor allele and HLA-B54:01 allele, resembling patterns observed in NAFLD. These results suggest that genetic variations in the HLA locus and their interplay with the gut microbiome may play a role in the development of NAFLD among lean individuals.
Discussion
This systematic review explored the genetic and epigenetic determinants of NAFLD among lean patients. To the best of our knowledge, this is the first and most updated systematic review to investigate genetic and epigenetic determinants of NAFLD among an understudied patient population, lean individuals. While NAFLD is commonly associated with obesity and metabolic syndrome, emerging evidence suggests that it can also affect individuals with a normal BMI. Understanding the genetic and epigenetic factors contributing to NAFLD in lean individuals is crucial, as it presents a unique subset of patients who exhibit the disease despite their lean phenotype. By exploring this distinct population, this study fills a critical knowledge gap and offers valuable insights into the complex etiology of NAFLD, potentially paving the way for personalized genetic and epigenetic diagnostic and therapeutic approaches tailored to lean individuals with this metabolic disorder.
The comprehensive systematic search process identified 18 studies exploring genetic and epigenetic determinants of NAFLD in lean individuals. These studies varied in design, sample size, and methodology, and were rigorously assessed for quality. Among the limited studies on epigenetic determinants, one study found a depletion of histone variants in the serum of lean NAFLD patients, suggesting their involvement in the disease’s pathogenesis. On the other hand, genetic determinants, particularly the PNPLA3 rs738409 G allele, were consistently associated with NAFLD risk in lean individuals across various populations. This allele was found to increase the risk of NAFLD, even in the absence of obesity and metabolic syndrome. Additionally, other genetic variants, such as the TM6SF2 rs58542926 allele, were associated with NAFLD susceptibility and serum triglyceride levels specifically in lean individuals. However, not all studies supported the presence of the PNPLA3 and TM6SF2 genetic associations, and contrasting findings were reported in some studies. Furthermore, the study highlighted the role of the CETP and APOC3 genes, as well as the HLA locus and gut microbiota, in the risk and development of NAFLD among lean individuals. These findings emphasize the complex interplay between epigenetic and genetic factors in lean NAFLD and underscore the need for further research to fully understand the mechanisms underlying the disease in this specific population.
Among the limited studies exploring epigenetic determinants in lean individuals with NAFLD, Buzova et al. conducted the only study specifically investigating this aspect, where they observed a depletion of histone variants in the serum of lean NAFLD patients, suggesting a potential epigenetic mechanism underlying the disease in this population (2). Their findings shed light on the potential involvement of histone modifications in the pathogenesis of NAFLD among lean individuals, contributing valuable insights into the epigenetic landscape of lean NAFLD. This is particularly necessary in the setting of the abundance of research on epigenetic determinants among non-lean NAFLD patients, which demonstrates that microRNAs and DNA methylation are elucidated as playing regulatory roles in NAFLD (20-23). For instance, Ahrens et al. reported nine enzymes (including PC, ACLY, PLCG1, IGF1, IGFBP2, and PRKCE) showing NAFLD-specific expression and methylation differences (21). Finally, it is important to note, however, that the literature on epigenetic determinants in lean NAFLD is considerably scarce compared to the extensive research on genetic factors, highlighting the need for further research in this area.
Furthermore, our results highlighted that the association between the PNPLA3 gene polymorphism and NAFLD risk in lean individuals has been extensively investigated, with a growing body of literature supporting its significance. This relationship has been consistently reported across diverse populations, including studies conducted in Hong Kong (11), Greece (13), Sri Lanka (6), Japan (4,5), Austria (7), and Italy (9). These studies have consistently shown that the PNPLA3 G allele is associated with an increased risk of NAFLD in lean individuals. Importantly, this association has also been observed in other populations, such as obese and non-normal BMI individuals. For instance, Wei et al. found a higher prevalence of the G allele among non-obese NAFLD patients compared to obese NAFLD patients in Hong Kong, suggesting that the PNPLA3 gene polymorphism contributes to NAFLD susceptibility beyond just lean individuals (11). These consistent findings across various populations highlight the robustness and generalizability of the association between the PNPLA3 gene polymorphism and NAFLD, indicating its crucial role in the pathogenesis of NAFLD, regardless of body weight or composition.
In lean individuals, the PNPLA3 GG polymorphism may increase their vulnerability to environmental factors, leading to the development of liver disease. Additionally, preliminary data suggests a significant association between the PNPLA3 GG polymorphism and NASH in patients with the lowest WC (9). Overall, recent studies in the literature have shown a strong connection between the PNPLA3 rs738409 gene polymorphism and increased liver fat content in individuals with NAFLD, irrespective of body weight or population type (24-27). As a result, the association between the PNPLA3 polymorphism and the development of NAFLD has become a prominent area of research interest (24).
It is important to note that there are few studies that have reported contrasting findings regarding genetic and clinical characteristics in this specific population. For instance, studies conducted by Zeng et al. and Hagström et al. did not find significant genetic associations in lean NAFLD patients (10,18). These studies suggest that factors other than the PNPLA3 gene may contribute to the development of NAFLD in lean individuals. Interestingly, Hagström et al. reported an increased risk of severe liver disease in lean NAFLD patients, despite the lack of significant genetic associations (10). These findings indicate that lean individuals with NAFLD may have distinct clinical characteristics and disease progression compared to their obese counterparts and remain understudied. Despite this, and while future research is required given the mixed results in the literature, the current evidence reveals that the PNPLA3 G allele is widely recognized as a risk allele for NAFLD among lean patients, but its impact may vary depending on the subpopulation and other factors.
Moreover, several studies have examined the relationship between TM6SF2 gene polymorphisms and the risk of NAFLD in lean individuals. These investigations have encompassed diverse populations, including Japanese (14), Indian (15), and multiethnic cohorts (17). Previous research in the literature has shown evidence in support of polymorphisms in TM6SF2 increasing risk for NAFLD and its association with more advanced fibrosis (28,29). Interestingly, the impact of TM6SF2 genetic variants on NAFLD susceptibility in lean subjects appears to be influenced by BMI. The findings suggest that certain TM6SF2 variants may confer a higher risk of NAFLD specifically in lean individuals, highlighting the complex interplay between genetic factors and BMI in the development of the disease. Furthermore, Hagström et al. reported an increased risk of severe liver disease among lean NAFLD patients, regardless of presence of any genetic risk allele (10). This underscores the clinical significance of studying these genetic associations to identify at-risk lean individuals. This is particularly important given that NAFLD is a common underlying liver disease in patients with hepatocellular carcinoma in the United States (30), and it is associated with type 2 diabetes (31), cardiovascular disease (32,33), and mortality (34,35). Furthermore, the severity of NAFLD has been shown to be driven, in part, by single-nucleotide polymorphisms in genes involved in lipid metabolism, oxidative stress, insulin signaling, and fibrogenesis (36). The clinical outcomes of non-obese or lean individuals with NAFLD compared with obese individuals with NAFLD remain unclear due to variation across recent studies (37,38). However, the presence of hepatic steatosis in individuals without a known cause of chronic liver disease, including excessive alcohol consumption, is the hallmark of NAFLD (39). Therefore, understanding the genetic associations of NAFLD is crucial for identifying individuals at risk of developing severe liver disease, and for developing targeted therapies to prevent or treat NAFLD and its associated complications.
In addition to the TM6SF2 gene, other genetic risk factors have been explored in lean individuals with NAFLD. Investigations have focused on CETP and APOC3 gene polymorphisms, shedding light on their potential roles in the development of the disease. Studies have revealed an association between CETP gene polymorphisms and NAFLD in females, suggesting a gender-specific effect. Furthermore, Asian Indian men carrying APOC3 variants have shown an increased prevalence of NAFLD. Notably, Yoshida et al. conducted a study specifically investigating the genetic determinants of NAFLD in lean Japanese patients. Their findings unveiled significant associations between the HLA locus, gut microbiota, and NAFLD among lean individuals (4).
Strengths and limitations
The present systematic review provides a comprehensive analysis of the genetic and epigenetic determinants NAFLD in lean individuals. One of the major strengths of this study is its pioneering nature, as it represents the first systematic review conducted specifically on this topic. By thoroughly reviewing the available literature, this study addresses a critical gap in the current knowledge regarding NAFLD, which is commonly associated with obesity but can also affect individuals with a lean body composition. Furthermore, the utilization of the NOS tool as a validated quality assessment method ensures that the included studies underwent rigorous evaluation, enhancing the reliability and validity of the findings presented in this manuscript.
Despite these strengths, it is important to acknowledge certain limitations of this review. Firstly, the predominance of cross-sectional study designs within the available literature restricts the ability to establish causation or determine temporal relationships between genetic and epigenetic factors and NAFLD in lean individuals. This limitation highlights the need for more longitudinal studies to elucidate the directionality and causal nature of these associations. Additionally, the absence of a meta-analysis due to the heterogeneity of the included studies limits the ability to provide quantitative estimations of effect sizes and statistical power. However, the qualitative synthesis conducted in this review provides a comprehensive narrative assessment of the available evidence. Moreover, the lack of a coherent and standardized definition of “lean” individuals across studies poses challenges in comparing and synthesizing the results, emphasizing the importance of establishing a consistent definition in future research. Furthermore, the potential for selection bias must be acknowledged, as the process of study selection may introduce biases. For instance, although efforts were made to conduct a comprehensive search, it is possible that some relevant studies were missed, especially those published in languages other than those included in the search or those not indexed in the searched databases. Additionally, the reliance on published studies introduces the risk of publication bias, where studies with statistically significant or positive results are more likely to be included, potentially overestimating treatment effects. Furthermore, time constraints inherent to the review process mean that new evidence generated after the search cutoff date may not be included, potentially impacting the comprehensiveness of the review. Moreover, inherent heterogeneity among the included studies, both in terms of study design and geographic representation, may limit the generalizability of the findings.
Finally, the limitations of the included studies must be acknowledged. The quality assessment of the included studies was conducted using NOS tool, as described in the methods section. The overall quality of the studies was determined to be moderate, with an average rating of approximately 8 stars out of 10. However, it is worth noting that several studies exhibited limitations in terms of comparability and ascertainments of exposure, indicating potential sources of bias. Notably, the studies demonstrated varying degrees of risk of bias.
Future research
The findings of this systematic review shed light on the genetic and epigenetic determinants of NAFLD in lean individuals, highlighting the importance of further research in this understudied population. Moving forward, several areas of future investigation can help deepen our understanding of the genetic and epigenetic determinants of NAFLD in lean individuals.
Firstly, longitudinal studies are needed to establish temporal relationships and elucidate the causal pathways between identified genetic and epigenetic factors and the development and progression of NAFLD in lean individuals. By assessing genetic and epigenetic profiles at baseline and following individuals over time, these studies can provide valuable insights into the predictive value of these factors and their impact on NAFLD outcomes and would also allow for causation to be established. Additionally, to ensure the generalizability of findings, future research should aim to include diverse populations in cohort studies. Currently, the literature on NAFLD in lean individuals is predominantly focused on specific ethnic and geographical populations. By expanding research efforts to include individuals from different ethnicities, geographic locations, and socioeconomic backgrounds, we can better understand potential variations in genetic and epigenetic determinants of NAFLD across populations. Additionally, such efforts can elucidate the role of age and ethnicity in relation to lean NAFLD, which can in turn direct further research and clinical guidelines.
Moreover, integration of multi-omics data holds great promise for advancing our understanding of NAFLD in lean individuals. Incorporating genomics, transcriptomics, epigenomics, and metabolomics data can provide a comprehensive view of the molecular underpinnings of NAFLD. This integrated approach will help identify novel biomarkers, unravel complex interactions between genetic and epigenetic factors, and facilitate the development of personalized prevention and treatment strategies. Furthermore, investigating the interplay between genetic/epigenetic determinants and lifestyle/environmental factors is crucial. Future research should explore the interactions between these factors and lifestyle-related factors such as diet, physical activity, gut microbiota, and environmental exposures. Understanding gene-environment interactions will contribute to a more comprehensive understanding of the risk factors for NAFLD among lean individuals and inform personalized prevention and intervention approaches.
Lastly, the development of predictive models that incorporate genetic and epigenetic markers, along with other clinical and lifestyle factors, can aid in identifying individuals at higher risk of developing NAFLD despite having a lean body composition. These models have the potential to improve early detection, risk stratification, and personalized management strategies for individuals at risk of NAFLD. Pursuing these research directions can bridge the knowledge gaps and further our understanding of the genetic and epigenetic determinants of NAFLD among lean individuals.
Conclusions
In conclusion, this systematic review highlights the importance of studying the genetic and epigenetic determinants of NAFLD among lean individuals, a population that presents a unique subset of patients exhibiting the disease despite their lean phenotype. The findings demonstrate the significant role of genetic factors, particularly the PNPLA3 rs738409 G allele, in NAFLD risk among lean individuals across various populations. Additionally, other genetic variants such as the TM6SF2 rs58542926 allele and epigenetic factors, including histone variants, show potential involvement in lean NAFLD. However, contrasting findings and the limited number of studies emphasize the need for further research. The review also underscores the clinical significance of these genetic associations, their impact on disease progression, and the potential for personalized diagnostic and therapeutic approaches. Future research should focus on longitudinal studies, diverse populations, multi-omics integration, gene-environment interactions, and predictive modeling to deepen our understanding of NAFLD in lean individuals and develop targeted interventions for this understudied population.
Acknowledgments
The authors would like to acknowledge all the researchers and authors whose valuable contributions and work in the field of NAFLD have made this systematic review possible.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-31/rc
Peer Review File: Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-31/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-31/coif). The authors have no conflicts of Interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Oxford: Ottawa Hospital Research Institute; 2000.
- Buzova D, Maugeri A, Liguori A, et al. Circulating histone signature of human lean metabolic-associated fatty liver disease (MAFLD). Clin Epigenetics 2020;12:126. [Crossref] [PubMed]
- Li J, Wu N, Yang Y, et al. Unique genetic variants of lean nonalcoholic fatty liver disease: a retrospective cohort study. BMC Endocr Disord 2023;23:11. [Crossref] [PubMed]
- Yoshida K, Yokota K, Kutsuwada Y, et al. Genome-Wide Association Study of Lean Nonalcoholic Fatty Liver Disease Suggests Human Leukocyte Antigen as a Novel Candidate Locus. Hepatol Commun 2020;4:1124-35. [Crossref] [PubMed]
- Honda Y, Yoneda M, Kessoku T, et al. Characteristics of non-obese non-alcoholic fatty liver disease: Effect of genetic and environmental factors. Hepatol Res 2016;46:1011-8. [Crossref] [PubMed]
- Niriella MA, Kasturiratne A, Pathmeswaran A, et al. Lean non-alcoholic fatty liver disease (lean NAFLD): characteristics, metabolic outcomes and risk factors from a 7-year prospective, community cohort study from Sri Lanka. Hepatol Int 2019;13:314-22. [Crossref] [PubMed]
- Feldman A, Eder SK, Felder TK, et al. Clinical and Metabolic Characterization of Lean Caucasian Subjects With Non-alcoholic Fatty Liver. Am J Gastroenterol 2017;112:102-10. [Crossref] [PubMed]
- Petersen KF, Dufour S, Hariri A, et al. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med 2010;362:1082-9. [Crossref] [PubMed]
- Fracanzani AL, Petta S, Lombardi R, et al. Liver and Cardiovascular Damage in Patients With Lean Nonalcoholic Fatty Liver Disease, and Association With Visceral Obesity. Clin Gastroenterol Hepatol 2017;15:1604-1611.e1. [Crossref] [PubMed]
- Hagström H, Nasr P, Ekstedt M, et al. Risk for development of severe liver disease in lean patients with nonalcoholic fatty liver disease: A long-term follow-up study. Hepatol Commun 2017;2:48-57. [Crossref] [PubMed]
- Wei JL, Leung JC, Loong TC, et al. Prevalence and Severity of Nonalcoholic Fatty Liver Disease in Non-Obese Patients: A Population Study Using Proton-Magnetic Resonance Spectroscopy. Am J Gastroenterol 2015;110:1306-14; quiz 1315. [Crossref] [PubMed]
- Shen J, Wong GL, Chan HL, et al. PNPLA3 gene polymorphism accounts for fatty liver in community subjects without metabolic syndrome. Aliment Pharmacol Ther 2014;39:532-9. [Crossref] [PubMed]
- Stasinou E, Argyraki M, Sotiriadou F, et al. Association between rs738409 and rs2896019 single-nucleotide polymorphisms of phospholipase domain-containing protein 3 and susceptibility to nonalcoholic fatty liver disease in Greek children and adolescents. Ann Gastroenterol 2022;35:297-306. [Crossref] [PubMed]
- Lin H, Wong GL, Whatling C, et al. Association of genetic variations with NAFLD in lean individuals. Liver Int 2022;42:149-60. [Crossref] [PubMed]
- Chahal D, Sharma D, Keshavarzi S, et al. Distinctive clinical and genetic features of lean vs overweight fatty liver disease using the UK Biobank. Hepatol Int 2022;16:325-36. [Crossref] [PubMed]
- Chatterjee A, Basu A, Das K, et al. Exome-wide scan identifies significant association of rs4788084 in IL27 promoter with increase in hepatic fat content among Indians. Gene 2021;775:145431. [Crossref] [PubMed]
- Stanislawski MA, Shaw J, Litkowski E, et al. Genetic Risk for Hepatic Fat among an Ethnically Diverse Cohort of Youth: The Exploring Perinatal Outcomes among Children Study. J Pediatr 2020;220:146-153.e2. [Crossref] [PubMed]
- Zeng J, Yang RX, Sun C, et al. Prevalence, clinical characteristics, risk factors, and indicators for lean Chinese adults with nonalcoholic fatty liver disease. World J Gastroenterol 2020;26:1792-804. [Crossref] [PubMed]
- Adams LA, Marsh JA, Ayonrinde OT, et al. Cholesteryl ester transfer protein gene polymorphisms increase the risk of fatty liver in females independent of adiposity. J Gastroenterol Hepatol 2012;27:1520-7. [Crossref] [PubMed]
- Xu Q, Li Y, Shang YF, et al. miRNA-103: molecular link between insulin resistance and nonalcoholic fatty liver disease. World J Gastroenterol 2015;21:511-6. [Crossref] [PubMed]
- Ahrens M, Ammerpohl O, von Schönfels W, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab 2013;18:296-302. [Crossref] [PubMed]
- Jin X, Feng CY, Xiang Z, et al. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget 2016;7:66455-67. [Crossref] [PubMed]
- Liu J, Lin B, Chen Z, et al. Identification of key pathways and genes in nonalcoholic fatty liver disease using bioinformatics analysis. Arch Med Sci 2020;16:374-85. [Crossref] [PubMed]
- Dai G, Liu P, Li X, et al. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity: A meta-analysis. Medicine (Baltimore) 2019;98:e14324. [Crossref] [PubMed]
- Byrne CD, Targher G. Liver fat content, non-alcoholic fatty liver disease, and risk of ischaemic heart disease. Eur Heart J 2018;39:3398. [Crossref] [PubMed]
- Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461-5. [Crossref] [PubMed]
- Mansoor S, Maheshwari A, Di Guglielmo M, et al. The PNPLA3 rs738409 Variant but not MBOAT7 rs641738 is a Risk Factor for Nonalcoholic Fatty Liver Disease in Obese U.S. Children of Hispanic Ethnicity. Pediatr Gastroenterol Hepatol Nutr 2021;24:455-69. [Crossref] [PubMed]
- Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352-6. [Crossref] [PubMed]
- Sookoian S, Castaño GO, Scian R, et al. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology 2015;61:515-25. [Crossref] [PubMed]
- Marrero JA, Fontana RJ, Su GL, et al. NAFLD may be a common underlying liver disease in patients with hepatocellular carcinoma in the United States. Hepatology 2002;36:1349-54. [Crossref] [PubMed]
- Ng CH, Chan KE, Chin YH, et al. The effect of diabetes and prediabetes on the prevalence, complications and mortality in nonalcoholic fatty liver disease. Clin Mol Hepatol 2022;28:565-74. [Crossref] [PubMed]
- Stols-Gonçalves D, Hovingh GK, Nieuwdorp M, et al. NAFLD and Atherosclerosis: Two Sides of the Same Dysmetabolic Coin? Trends Endocrinol Metab 2019;30:891-902. [Crossref] [PubMed]
- Shabalala SC, Dludla PV, Mabasa L, et al. The effect of adiponectin in the pathogenesis of non-alcoholic fatty liver disease (NAFLD) and the potential role of polyphenols in the modulation of adiponectin signaling. Biomed Pharmacother 2020;131:110785. [Crossref] [PubMed]
- Byrne CD, Targher G. Non-alcoholic fatty liver disease is a risk factor for cardiovascular and cardiac diseases: further evidence that a holistic approach to treatment is needed. Gut 2022;71:1695-6. [Crossref] [PubMed]
- Chang Y, Ryu S, Sung KC, et al. Alcoholic and non-alcoholic fatty liver disease and associations with coronary artery calcification: evidence from the Kangbuk Samsung Health Study. Gut 2019;68:1667-75. [Crossref] [PubMed]
- Sookoian S, Pirola CJ. Genetic predisposition in nonalcoholic fatty liver disease. Clin Mol Hepatol 2017;23:1-12. [Crossref] [PubMed]
- Chun HS, Lee M. Lean vs. obese phenotypes of nonalcoholic fatty liver disease: similar or different? Clin Mol Hepatol 2023;29:377-80. [Crossref] [PubMed]
- Tan EXX, Muthiah MD, Ng CH, et al. Editorial: clinical outcomes in lean NAFLD-the devil is in the details. Aliment Pharmacol Ther 2023;57:1040-1. [Crossref] [PubMed]
- Younossi ZM, Stepanova M, Negro F, et al. Nonalcoholic fatty liver disease in lean individuals in the United States. Medicine (Baltimore) 2012;91:319-27. [Crossref] [PubMed]
Cite this article as: Njei B, Al-Ajlouni YA, Ugwendum D, Abdu M, Forjindam A, Mohamed MF. Genetic and epigenetic determinants of non-alcoholic fatty liver disease (NAFLD) in lean individuals: a systematic review. Transl Gastroenterol Hepatol 2024;9:11.