
Analysis of prognostic germline polymorphisms in patients with advanced hepatocellular carcinoma
Highlight box
Key findings
• Single nucleotide polymorphisms (SNPs) prognostic in advanced hepatocellular carcinoma (HCC) were not validated in our dataset. Novel immunomodulatory SNPs in CCL2, IL-10, cytotoxic T-lymphocyte antigen-4 (CTLA-4) and NFKB1 were however found to be prognostic.
What is known and what is new?
• Sorafenib remains a key therapy for advanced HCC, despite recent novel treatment options. Observational data has suggested SNPs involved in genes in angiogenesis including VEGF, eNOS, ANGPT2 and VEGFR2 are prognostic in HCC.
• Prognostic SNPs from the literature were not externally validated. Exploratory SNPs within immunomodulatory pathways were significantly associated with prognosis in advanced HCC.
What is the implication, and what should change now?
• Identified prognostic immunomodulatory SNPs should be externally validated, and could provide guidance for treating clinicians and new targets for intervention.
Introduction
Background
Hepatocellular carcinoma (HCC) is the 7th most common cause of cancer, and the 2nd leading cause of cancer deaths worldwide (1). The most important risk factor for HCC is liver cirrhosis, most often related to viral hepatitis. Additional risk factors include alcohol use, smoking, non-alcoholic fatty liver disease (NAFLD), and others.
HCC is a highly vascular tumor, underlining the importance of angiogenesis in disease progression. Research has shown angiogenesis in HCC is stimulated by direct production of angiogenic factors such as vascular endothelial growth factor (VEGF), angiopoietin-2 (Ang2), platelet-derived growth factor (PDGF) by malignant cells as well as infiltrating inflammatory cells (2). This occurs both due to hypoxia at the centre of the tumor, and through a constitutive production of angiogenic factors caused by mutations in tumor suppressor and oncogenes (2). High circulating levels of VEGF and high tumor microvessel density are associated with a more aggressive clinical course in HCC and worse survival (3).
The HCC microenvironment is composed of multiple cell types with immunomodulatory properties that can support carcinogenesis. A complex interplay between malignant cells and immune cells in the tumor microenvironment (TME) involving both immune stimulatory and immunosuppressive factors ultimately result in a milieu that prevents eradication of the tumor by the immune system. This signaling is mediated by a diverse set of molecules and cytokines (4) (Figure 1).
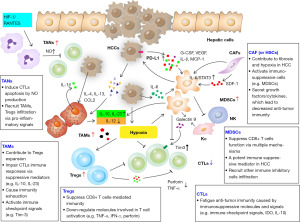
Multiple lines of research suggest crosstalk between angiogenesis and regulation of the immune microenvironment. Pro-angiogenic factors have a role in promoting an intratumoral immunosuppressive microenvironment. Signaling through VEGFR expressed on dendritic cells (DCs) has been shown to inhibit DC maturation (5). VEGF can promote the accumulation of immunosuppressive myeloid derived suppressor cells (MDSCs) in the TME (6). Moreover, VEGF can induce proliferation of other immunosuppressive cell types including Tregs (7) and tumor associated macrophages (TAMs). There is also evidence this relationship between angiogenesis and the immune system is reciprocal, with many immune cells fostering a niche that is supportive of angiogenesis. Both innate and adaptive immune cells within the TME can develop phenotypes that promote angiogenesis through several mechanisms. TAMs can support angiogenesis through the release of proangiogenic factors including VEGF and fibroblast growth factor-2 (FGF2) (8) as well as through breakdown of the extracellular matrix via the production of matrix metalloproteinases (MMPs) (9). Studies of immature myeloid cells including MDSC and DCs have demonstrated proangiogenic properties in the TME including the ability to release angiogenic factors such as VEGF as well as the ability to transform into TAM’s or even endothelial cells (10). A recent successful clinical trial combined an anti-angiogenic agent (bevacizumab) with immune checkpoint inhibition (atezolizumab) with improvement in overall survival (OS), suggesting synergy between these pathways (11).
Sorafenib was the first systemic therapy to show convincing improvement of OS in advanced HCC in large, randomized trials. As a result of the landmark SHARP and Asia Pacific trials, sorafenib was widely adopted for the first line treatment of advanced HCC, however overall response rate (ORR) was low at 2%, and OS only improved from 7.9 to 10.7 months (12). Sorafenib is an orally administered, multi-serine/threonine and tyrosine kinase inhibitor (TKI) that targets VEGFR 1–3, PDGFR, FGFR1, KIT, RET, FLT3 and RAF pathways (13). The anticancer effects of sorafenib are thought to result both from inhibition of growth promoting intracellular signaling and from disruption of angiogenesis signaling in the TME. Only recently have additional systemic therapies shown convincing evidence of efficacy in advanced HCC, including additional oral TKIs, and atezolizumab, a programmed death-ligand 1 (PD-L1) inhibitor, in combination with bevacizumab. Despite these new options, many patients around the world will still receive treatment with sorafenib in a later line of therapy, or because of contraindications to atezolizumab or bevacizumab or because the high costs of these therapies make them unaffordable to most patients.
Rationale and knowledge gap
The prognosis of patients with HCC can be affected by several tumor specific and patient related factors. A systematic review by Tandon et al. identified 72 studies examining 79 prognostic factors in HCC, with almost 24,000 patients included. Five most common independent predictors of mortality were portal vein thrombosis, tumor size, Child-Pugh class, bilirubin and alpha-fetoprotein (AFP) (14). Validated prognostic biomarkers in HCC are lacking.
Single nucleotide polymorphisms (SNPs) are an attractive biomarker class due to their ease of measurement and ability to effect both tumor and host biology. In the context of cancer, SNPs have been most widely studied as risk factors for cancer development. In the context of HCC, Penha Mesquita et al. performed a systematic search of meta-analyses with revaluation by Bayesian calculations and found polymorphisms in CCND1, CTLA4, EGF, IL6, IL12A, KIF1B, MDM2, MICA, miR-499, MTHFR, PNPLA3, STAT4, TM6SF2 and XPD to be indicative of HCC risk (15). There are, however relatively sparse data existing on how SNPs influence prognosis in patients with existing cancer diagnoses. Polygenic risk scores are not currently used to inform prognosis in patients with established cancer diagnoses. There are however examples of individual SNPs found to have prognostic or predictive relevance for patient outcomes include CYP3A4 in prostate cancer treated with androgen deprivation therapy, and TPMT in childhood leukemia receiving 6-mercaptopurine based treatment (16).
Objective
We aimed to review the literature to identify biomarkers with prognostic value in patients with advanced HCC, with a focus on SNPs. Patients with advanced HCC treated with Sorafenib were the main patient population of interest. Sorafenib was the standard of care therapy for advanced HCC at the time of study planning. From this initial literature search we aim to select SNPs with evidence for use as prognostic biomarkers, and externally validate them using a retrospective cohort of patients with advanced HCC at our institution. Furthermore, we will test exploratory immunomodulatory SNPs in our patient cohort. We aimed to analyze these SNPs in TACE treated patients because they also represent a group with incurable HCC. This allows for hypothesis generation as to whether the analyzed SNPs are prognostic in advanced HCC or more specifically predictive of sorafenib effect. We present this article in accordance with the STROBE reporting checklist (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-22/rc).
Methods
Structured literature review
A structured literature review of MEDLINE including Epub ahead of print, and EMBASE was performed to identify existing SNPs found to be prognostic in sorafenib treated patients with HCC. Refer to the search strategy document for details on the search algorithm.
Titles of potentially relevant studies results were selected from the list of search results. Following this, abstracts were reviewed for studies meeting eligibility criteria. The references of included studies were reviewed to find additional publications for inclusion. Studies identified in the structured review are listed (Table S1). The magnitude of the effect size of prognostic biomarkers was reported where available.
Eligibility criteria for study selection includes:
- Studies must have included patients with advanced/incurable HCC, who were treated with sorafenib;
- Included patients 18 years or older;
- Studies must have evaluated the prognostic or predictive ability of one or more SNPs against the clinical endpoints of either ORR, disease control rate, progression-free survival (PFS) or OS and have demonstrated a statistically significant association;
- Published in the English language.
From this literature review, manuscripts of publications evaluating SNPs as prognostic biomarkers in sorafenib treated patients with HCC were reviewed. To avoid multiple comparisons, a maximum of 8 SNPs of interest were selected for external validation. Only SNPs with statistically significant findings in the original article were chosen. The disposition of studies identified in the literature review is presented in Figure S1. Additional factors forming the basis of SNP selection included strength of association [hazard ratio (HR), odds ratio (OR), relative risk (RR)], replication of SNP in multiple studies, biological plausibility, and study quality. Study quality was performed by assigning each manuscript a score using the REMARK framework (17). SNPs from studies with inadequate quality using the REMARK framework were not included for external validation.
Analysis of exploratory prognostic SNPs in patients with advanced HCC
Exploratory SNPs were selected using a candidate gene approach. Candidate genes were chosen that have a potential functional impact on either immunomodulation in the HCC TME, or impact on regulation of the immune system angiogenesis interface. To achieve this, a literature review was conducted to identify the immune cell types present in the HCC TME. Further, the literature was reviewed of cell types and signaling pathways involved in the interface between the immune system and angiogenesis. The full list of included cell type is as follows: TAMS, tumor associated neutrophils (TANs), mast cells, eosinophils, MDSCs, natural killer (NK), natural killer T cells (NKT), DCs, cytotoxic T lymphocytes (CTLs), Tregs, Kupffer, and endothelial cells. Genes critical to the regulation of the above cell types, or genes encoding the signaling networks between the above cell types were chosen. Using this approach, the full list of analyzed genes includes: ICAM1, VCAM1, EDNRA/B, EMAP2, Ang2, Tie2, IL1B, IL-A, IL2, IL4, IL5, IL6, IL8, IL10, IL12, IL13, IL17, IL18, M-CSF, CSFR1, MCP-1, SDF-1, SEMA3A, NRP1, GCSF, GM-CSF, STAT3, NFKB, IFNa, TNFa, TGFB, OncostatinM, CCL2, CCL3, CCL4, CCL5, CCL11, CCL15, CCL22, CCL28, CCR2, CXCL1, CXCL2, CXCL3, CXCL4, CXCL5, CXCL8, CXCL9, CXCL10, CXCL12, CXCL17, CXCR3, CXCR4, Bv8, iNOS, ARG1, IRF8, MMP1, MMP7, MMP9, MMP12, PD-L1, PD-1, CTLA-4, CD80, TIM3, LAG3, ICOS, FOXP3, GITR, Galectin9, CD25.
From this list of candidate genes, another literature review was conducted to identify SNPs with a high level of evidence as being functionally active in the regulation of inflammation or angiogenesis. To achieve this, a search of OMIM and PubMed was conducted including each gene name, and the terms “SNP” and either “inflammation” or “angiogenesis”. SNPs showing a statistically significant association with either laboratory or clinical markers of inflammation or angiogenesis in at least two independent studies were chosen for inclusion in the exploratory analysis.
This final list of exploratory SNPs can be found in the Table S2 and includes: rs10204525, rs1024611, rs1036199, rs1143627, rs1143634, rs11568818, rs11568821, rs16944, rs17561, rs17576, rs1799750, rs1799969, rs1799983, rs1800469, rs1800587, rs1800629, rs1800795, rs1800872, rs1800896, rs1870377, rs2010963, rs20541, rs2069762, rs2070744, rs2070874, rs2071559, rs2227306, rs2232365, rs2243250, rs2275913, rs2276109, rs2297518, rs231775, rs28362491, rs3024505, rs3212227, rs3761548, rs3816769, rs4073, rs4359426, rs4604006, rs5498, rs55633437.
Study patient population
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Princess Margaret Cancer Centre (No. IRB00001258) and individual consent for this retrospective analysis was waived.
Patients for this study were drawn from a larger cohort of patients recruited to the Molecular Epidemiology of Hepatobiliary Tumors (HBT) Study at our institution. The population of interest included patients over the age of 18 with advanced HCC treated at our institution within the HBT database who have stored blood samples. One cohort included patients treated with sorafenib between October 1st 2008 and November 1st 2019, and another cohort included patients who received trans arterial chemoembolization (TACE) between January 1st 2002 and November 1st 2019. Starting dates reflect data availability in the HBT database. Patients must have had a diagnosis of HCC either through histologic confirmation or clinical diagnosis according to American Association for the Study of Liver Diseases (AASLD) guidelines (18). The decision for treatment was made by the treating clinician.
In the sorafenib cohort patients were selected with advanced HCC [Barcelona-Clinic Liver Cancer (BCLC)-C] or intermediate stage HCC (BCLC-B) refractory to or ineligible for curative therapies or chemoembolization. All patients received sorafenib at a dose of 400 mg po bid continuously with dose reductions applied as clinically indicated. In the TACE cohort patients were selected with intermediate stage HCC (BCLC-B), or early-stage HCC (BCLC-A) refractory to or ineligible for curative therapies alone. Patients with portal vein thrombus who were reviewed in a multidisciplinary conference and felt to be eligible for TACE were included. All patients received TACE with chemotherapeutic agents including doxorubicin, cisplatin or mitomycin, which were mixed with lipiodol, and emulsified with water-soluble contrast. Patient cohort details are outlined in the Consort diagram (Figure 2).
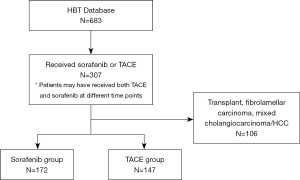
Additional eligibility criteria as follows:
Inclusion criteria:
- Banked blood sample available for SNP testing;
- Age 18 years or older;
- Date of blood collection January 1 2010–present;
- Received one of the following treatment modalities for HCC:
- TACE;
- Sorafenib.
Exclusion criteria:
- Mixed HCC/cholangiocarcinoma;
- Fibrolamellar HCC;
- Patients undergoing liver transplantation during treatment course.
Data collection
Clinical data was collected from electronic health records. Collected variables included demographic data, risk factors for HCC such as hepatitis virus infection status, prognostic variables, treatment data and outcomes. Outcome measures collected included date of disease progression, date of death, and requirement for dose reductions. The list and definition of variables can be found in the data management document. Data are stored in password protected files. Existing data were utilized from previously collected sources including existing databases. For these patients, data quality was assessed by randomly selecting 10% of cases for secondary review.
Performance criteria
Selected biomarkers were evaluated on their prognostic value for patient outcomes.
Primary outcome:
- OS: defined as time from treatment (either TACE, or start of sorafenib) to death or censorship.
Secondary outcome:
- Real world PFS: defined as time from TACE or sorafenib until radiologic or physician-assessed disease progression or death. PFS can be a surrogate for OS, is clinically relevant to patients and often has improved power.
For patients with more than one treatment course of TACE, the first of such treatment was used for analysis of outcomes. For patients treated with multiple different treatment modalities throughout their illness course, outcomes were analyzed for each respective treatment modality (i.e., TACE or sorafenib).
Statistical analysis
Baseline characteristics were reported as medians and ranges for continuous variables and frequencies with percentages for categorical variables. Departure from Hardy-Weinberg equilibrium (HWE) were tested using the Pearson χ2 test with a significant departure from HWE consider as P=5e−05. Linkage disequilibrium (LD) between SNPs were calculated as D’ using the genetics package in RStudio.
Time-to-event data, such as PFS, were summarized using the Kaplan-Meier method using the starting date of sorafenib (or TACE) to the date of progression, death or last follow up. OS was estimated using the Kaplan-Meier method using the starting date of treatment to the date of death or last follow up.
The log-rank test was used to test univariable associations between clinical factors and OS, PFS. Necessary transformation of continuous variables was tested by examining martingale residuals. Complete case analysis was used to deal with missing data. Genotypes were coded using the additive model. A base clinical model was built to determine the independent predictive value of sequence variants. The base model included known prognostic factors determined from literature review including Child-Pugh score, T-stage, portal vein thrombus, AFP and neutrophil-lymphocyte ratio (NLR). Additional clinical factors were chosen via backward selection to retain in the clinical model with a significance level for staying (SLS) in the model cut-off of 0.2. SNPs were tested for prognostic ability using a log-rank test as well as likelihood ratio tests when added to the base clinical model. P values, HR and their 95% confidence intervals (CIs) for survival were reported. A two-sided type I error rate of 5% was used for testing in this setting. The detectable HR for OS, assuming 80% statistical power and a two-sided significance level at 0.05, a sample size of 152 patients with an event rate of 106 (70% of patients) and the proportion of patients with a SNP of interest being 10%, is 0.327. A multivariable model including the base clinical variables and SNPs found to be significant on univariable testing was created. The proportional hazards assumption was tested for variables using Schonenfeld residuals. Statistical tests will be performed using R software.
Blood specimen analysis
Up to two tablespoons of blood was taken (30 cc or less) in purple top EDTA (whole blood), red top (serum), and yellow top Na-heparin (plasma) tubes. A research/laboratory technician stored the samples in a −70 ℃ freezer. Each peripheral blood sample DNA was extracted by ROCHE DNA Isolation Kit for Mammalian Blood (cat#11667327001). DNA yield and quality was controlled by Nanodrop. All the DNA samples were plated on four 96-well plates in 50 ng/µL, 10 µL well/sample. Sample plates were submitted to TCAG (https://www.tcag.ca) for MassARRAY genotype.
Samples were genotyped for 43 SNP positions in 2 (multiplex) panels using the MassARRAY® Analyzer 4 System (Agena Biosciences, San Diego, CA, USA) using iPlex Pro chemistry and analyzed using Typer 4.0 software. Briefly, each locus is amplified by polymerase chain reaction (PCR) and a third primer that flanks the polymorphism site is extended by one base (primer sequences are available upon request). The extension reaction products are analyzed using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry and correlated (by size) to the SNV and nucleotide observed. Commercial reference samples NA12878, NA12891, NA12892, NA18942 and NA18947 were genotyped with the study samples to validate each assay. Genotyping was performed blinded to outcomes.
Results
SNP genotyping results
Descriptions of polymorphisms selected for the external validation cohort and exploratory cohort are displayed in the Table S3. Genotypes are reported using the short form A (adenosine), C (cytosine), T (thymine), or G (guanine). All SNPs genotyped had an overall call rate >98%, except for one SNP rs1143627 which failed genotyping and was not included in the analysis (Table S4) for sample genotype and allele frequency. General population allele frequency was accessed via ALFA from dbSNP. Allele frequency of study participants was similar to the general population. All SNPs except for IL-4 (rs2070874), FOXP3 (rs2232365), IL-4 (rs2243250), FOXP3 (rs3761548) were in Hardy Weinberg equilibrium.
Patient characteristics-sorafenib treated HCC group
In total, 172 patients with HCC that were treated with sorafenib were included for analysis (Table 1). The median follow up was 10.6 months. The median age was 66.5 years, and 83% were male. The most common ethnicities were Caucasian and Asian. The etiology of cirrhosis in patients was hepatitis C virus (HCV) (36%), hepatitis B virus (HBV) (27%), alcohol (19%), and NAFLD (13%). Patients had frequently received prior therapy for HCC including resection, radiofrequency ablation (RFA), radiation and TACE. About 84% had multifocal HCC and half had portal venous tumor thrombus, half of patients had extrahepatic spread of tumor. And 93% had Child-Pugh A or B liver function. Dose reductions during sorafenib treatment were frequent at 80%. There were 106 deaths during follow-up. The median OS of all sorafenib treated patients was 15.4 months. There were 156 patients with progression or death during follow up, and the median PFS was 5.2 months.
Table 1
| Clinical characteristic | Value (N=172) |
|---|---|
| Gender | |
| Male | 143 [83] |
| Female | 29 [17] |
| Age (years) | 66.5 (17.6–87.2) |
| Ethnicity | |
| Asian/pacific islander | 59 [34] |
| Caucasian | 85 [49] |
| Black | 4 [2] |
| Latino | 1 [1] |
| Aboriginal | 0 |
| Other/mixed | 10 [5] |
| Missing | 13 [8] |
| Etiology | |
| HBV | 44 [27] |
| HCV | 61 [36] |
| Alcohol | 32 [19] |
| NAFLD | 22 [13] |
| Other | 13 [5] |
| BCLC | |
| A | 4 [2] |
| B | 20 [12] |
| C | 145 [84] |
| Missing | 3 [2] |
| Serum AFP (ng/mL) | |
| ≥200 | 77 |
| <200 | 86 |
| Missing | 9 |
| Prior therapy | |
| Surgical resection | 30 [17] |
| RFA | 52 [30] |
| TACE | 58 [34] |
| Radiation | 50 [29] |
| Transplant | 0 |
| Multifocal | |
| Yes | 144 [84] |
| No | 28 [16] |
| PVT | |
| Yes | 88 [51] |
| No | 84 [49] |
| Child-Pugh score | |
| A5 | 118 [69] |
| A6 | 41 [24] |
| B7 | 10 [6] |
| ≥B8 | 3 [1] |
| Extrahepatic disease | |
| Yes | 91 [53] |
| No | 81 [47] |
| NLR* | |
| ≥3 | 94 |
| <3 | 71 |
| Missing | 7 |
| Dose reduction of sorafenib | |
| Yes | 138 [80] |
| No | 34 [20] |
Data are presented as n [%] or median (range). *, optimal cut-off based on the review by (19). HBV, hepatitis B virus; HCV, hepatitis C virus; NAFLD, non-alcoholic fatty liver disease; BCLC, Barcelona-Clinic Liver Cancer; AFP, alpha-fetoprotein; RFA, radiofrequency ablation; TACE, trans arterial chemoembolization; PVT, portal venous thrombosis; NLR, neutrophil-lymphocyte ratio.
Univariable analysis
Clinical variables
Univariable testing of clinical variables revealed a statistically significant association of only the Child-Pugh score with OS (median survival 19.2 months for Child-Pugh 5, 9.6 months for Child-Pugh 6, 11.9 months for Child-Pugh 7) and no clinical variables were significant predictors of PFS (Table S5).
Validation SNPs genotype analysis
Univariable analysis of SNPs selected for validation revealed only VEGFR2 (rs1870377) [minor allele frequency (MAF) =0.24] was statistically significant for prognosis of PFS (median 2.9 months AA, 5.3 months AT, 5.3 months TT, P=0.036 on log-rank testing, P=0.048 on nested likelihood ratio, HR 1.35 95% CI: 1.006–1.822), but not for OS (Figure 3, Table 2). No other SNPs in the validation cohort were significant for PFS or OS.
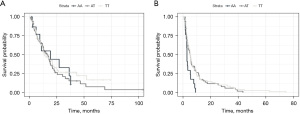
Table 2
| Gene | Genotypes (%) | Median PFS (months) | Median OS (months) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wt. | Het. | Homo. | P | Wt. | Het. | Homo. | P | |||
| VEGFR2 rs1870377 | AA/AT/TT (11/38/51) | 5.3 | 5.3 | 2.9 | *0.036, +0.048 | 13.7 | 16.1 | 19.2 | *0.6, +0.61 | |
*, log rank statistical test; +, nested LR statistical test. SNPs, single nucleotide polymorphisms; PFS, progression-free survival; OS, overall survival; wt, wild type; Het, heterozygous; Homo, homozygous; LR, likelihood ratio.
Exploratory SNPs genotype analysis
Univariable analysis of exploratory genotypes revealed that CCL2 (rs1024611) (MAF =0.28) was a significant predictor of prognosis for OS (12.0 months AA, 20.8 months AG, 21.1 months GG, P=0.004) but not for PFS. The SNP IL-10 (rs1800896) (MAF =0.45) was prognostic for PFS (9.6 months CC, 5.2 months CT, 4.8 months TT, P=0.0047) and for OS (25.5 months CC, 14.9 months CT, 13.0 months TT, P=0.014). CTLA-4 (rs231775) (MAF =0.37) was prognostic for PFS (5.5 months AA, 5.3 months AG, 3.0 months GG, P=0.035) but not for OS. NFKB1 (rs28362491) (MAF =0.42) was a predictor of PFS (5.7 months ATTG/ATTG, 5.1 months ATTG/DEL 3.6 months DEL/DEL, P=0.0087) and for OS (20.8 months, 12.9 months, 13.4 months, P=0.047) (Figure 4, Table 3).
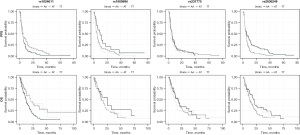
Table 3
| Gene | Genotypes (%) | Median PFS (months) | Median OS (months) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wt. | Het. | Homo. | P | Wt. | Het. | Homo. | P | |||
| CCL2 rs1024611 | AA/AG/GG (49/35/16) | 5.0 | 5.3 | 3.8 | *0.3, +0.47 | 12.0 | 20.8 | 21.1 | *0.004, +0.065 | |
| IL-10 rs1800896 | CC/CT/TT (12/39/49) | 4.8 | 5.2 | 9.6 | *0.02, +0.0047 | 13.0 | 14.9 | 25.5 | *0.3, +0.014 | |
| CTLA4 rs231775 | AA/AG/GG (31/49/20) | 5.5 | 5.3 | 3.0 | *0.2, +0.035 | 17.6 | 18.1 | 10.9 | *0.4, +0.37 | |
| NFKB1 rs28362491 | ATTG.ATTG/ATTG.DEL/DEL.DEL (40/46/15) | 5.7 | 5.1 | 3.6 | *0.01, +0.0087 | 20.8 | 12.9 | 13.4 | *0.3, +0.047 | |
*, log rank statistical test; +, nested LR statistical test. SNPs, single nucleotide polymorphisms; PFS, progression-free survival; OS, overall survival; wt, wild type; Het, heterozygous; Homo, homozygous; LR, likelihood ratio.
Multivariable analysis SNPs sorafenib group
The clinical variables that were specified a-priori for inclusion in the multivariate model were gender, Child-Pugh score, T stage, portal vein tumor thrombus, AFP level and NLR. SNPs with a significant association on univariable testing were also included in the multivariable model with a final list of: VEGFR2 (rs1870377), CCL2 (rs1024611), IL-10 (rs1800896), CTLA4 (rs231775), and NFKB1 (rs28362491). Significant predictors of OS included T-stage, CCL2 (rs1024611), and IL-10 (rs1800896). Significant predictors of PFS included NFKB1 (rs28362491) (Table S6). None of the SNPs in the validation cohort were significant predictors of PFS or OS in the multivariate model. Testing of the proportional hazards assumption using Schoenfeld residuals revealed AFP was the only variable that violated the PH assumption.
TACE treated HCC group analysis
In total 147 patients with HCC that were treated with TACE were included for analysis (see Table S7 for description of clinical characteristics). The median survival of patients in the TACE database was 26.2 months, and the median PFS was 4.5 months.
Univariable analysis of clinical variables in the TACE treated cohort revealed an association of pre-treatment AFP level with OS. There was a significant association between age and BCLC stage with PFS (Table S8).
Univariable analysis SNPs-TACE Group
Among SNPs in the validation cohort, there was an association between NOS3 (rs2070744) and PFS (C/C 9 months, C/T 4.4 months, T/T 4 months, P=0.039), but not OS. There was an association with VEGFR2 (rs1870377) and OS on log rank testing but not on likelihood ratio testing compared to the base clinical model. Similarly, there was an association between CCL2 (rs1024611) and PFS on log rank testing but not on likelihood ratio testing compared to the base clinical model. TNF (rs1800629) was associated with OS (A/A 41 months, A/G 45 months, G/G 23.9 months P=0.007) but not PFS. IL-13 (rs20541) was associated with OS (G/G 25.5 months, A/G 30.7 months, A/A 45 months P=0.048) but not PFS. Finally, NFKB1 (rs28362491) was associated with OS (ATTG/ATTG 25.8 months, ATTG/DEL 24.3 months, DEL/DEL 28.8 months P=0.04 (Table S9).
Discussion
Key findings
Sorafenib remains an important treatment in a growing armamentarium for advanced HCC. However, response rates and improvements in survival are modest so selecting patients that may benefit would aid the decision making of clinicians. Prognostic factors used in the clinical setting have been incorporated into staging systems and include tumor size and spread, portal venous involvement, markers of liver function and patient functional status. Furthermore, AFP and NLR values have robust evidence for prognosis.
Thus far there have been limited clinically useful biomarkers for prognosis in patients with advanced HCC beyond those above mentioned (Table S1). SNPs are an attractive class of biomarkers as they do not vary with time and are readily measurable from blood samples. Prognostically relevant SNPs may help stratify poorer risk patients, avoid over treatment, unnecessary side effects or help in determining more appropriate treatment strategies. Examples of SNPs with clinical utility in cancer treatment include the association of XRCC1 codon 399 with prediction of platinum benefit in gastrointestinal, genitourinary and lung cancer. XRCC1 is involved in DNA repair, a major therapeutic mechanism of platinum agents (20). In advanced prostate cancer, polymorphisms in CYP19A1 effect testosterone metabolism and are associated with prognosis in androgen deprivation therapy treated patients (21).
We performed a literature review of studies examining the prognostic implications of SNPs in patients receiving sorafenib for advanced HCC and found data that the following SNPs were prognostic for clinically relevant outcomes: VEGF-C rs4604006, VEGF-A rs2010963, eNOS rs2070744, eNOS rs1799983, ANGPT2 rs55633437, VEGFR2 rs1870377, and VEGFR2 rs2071559.
In the ePHAS study (22), 128 Italian patients were retrospectively analyzed. Included patients had advanced HCC and had received treatment with sorafenib. Polymorphisms of eNOS were analyzed for prognostic value under the hypothesis that sorafenib, by inhibiting VEGF-R would reduce nitric oxide production. Three eNOS (NOS3) polymorphisms were analyzed including −786T>C (rs2070744), VNTR 27bp 4a/b and +894G>T (rs1799983). Linkage disequilibrium was observed between eNOS-786 and eNOS VNTR (D’=0.85), and 4 haplotypes were observed (HT1-4). In the validation cohort, multivariate analysis revealed significant associations of eNOS-786 with PFS (HR 5.87), eNOS VNTR with OS (HR 7.04), eNOS+894 with OS (HR 11.95), and HT1 with PFS and OS (HR 11.17 and 7.03 respectively).
Marisi et al. (23), retrospectively studied 135 Italian patients with advanced HCC receiving sorafenib. Polymorphisms in ANGPT2, a signaling molecule critical in angiogenesis were tested for prognostic significance, as were the eNOS polymorphisms previously described in the ePHAS study. Eight ANGPT2 SNPs were selected for inclusion. In multivariate analysis, ANGPT2 rs55633437 and NOS3 rs2070744 were found to be independent prognostic factors predicting PFS (HR 0.24; HR 6.32, respectively) and OS (HR 0.67; HR 5.48, respectively).
ALICE-1 (24) was a retrospective multicentre study that evaluated 148 patients with intermediate to advanced HCC who received treatment with sorafenib. The aim of the study was to assess SNPs in the VEGF and VEGFR genes to determine if they were prognostic in this group of patients. On multivariate analysis, rs2010963 (VEGF‐A), rs4604006 (VEGF‐C) were found to be independent prognostic factors for PFS (HR 0.25, 0.22 respectively) and OS (HR 0.28, 0.25 respectively).
A retrospective study was conducted by Zheng et al. (25) of 78 patients with advanced HCC treated with sorafenib in China. TagSNPs of VEGFR2 with a MAF >0.1 were chosen by analysis of HapMap genotyping data. Four SNPs with that were nonsynonymous and another SNP in the promoter region of VEGFR2 previously found to be functional were also included. On multivariate analysis, SNPs that were found to be prognostic included rs1870377 [time to progression (TTP): HR 0.68, OS: HR 0.35], and rs2071559 (OS: HR 2.25).
REMARK framework scoring demonstrated that each study had limited methodological deficiencies but were of an acceptable quality that the results could be tested as to their external validity (Appendices 1-4).
We aimed to external validate this list of SNPs to determine if these results were reproducible in our database of patients with advanced HCC. Furthermore, we aimed to analyze novel SNPs within immunomodulatory pathways to determine their prognostic significance in advanced HCC.
We failed to validate existing prognostic SNPs from the literature in our dataset. We did find that one SNP, rs1870377 was significantly associated with outcome in our dataset, however the direction of effect was opposite to that in the source paper. We did find that the immunomodulatory SNPs CCL2 (rs1024611), IL-10 (rs1800896), CTLA-4 (rs231775) and NFKB1 (rs28362491) were prognostic in our dataset.
Strength and limitations
Sorafenib was the only approved therapy for advanced HCC during the dates of data collection for this study, and therefore is the agent all patients would have received if not deemed ineligible or if patients opted to pursue palliative care alone. In order to determine whether candidate biomarkers are predictive of sorafenib response instead of simply prognostic, they must be studied in a cohort of patients with HCC who are not exposed to sorafenib, which would not be possible by reviewing retrospective patient data. As an exploratory analysis, we evaluated candidate biomarkers in patients with HCC treated with TACE to determine whether they might be prognostic in this group also. An exploratory analysis of the interaction between biomarker status and treatment modality will be performed to determine whether tested biomarkers gave different results in patients not treated with sorafenib. This is a limitation because patients in the control group will have earlier stage, lower volume disease with a better prognosis and so comparisons will be indirect, although these potential confounders will be included in the statistical model to control for this where possible.
Comparison with similar research and explanation of findings
Of the SNPs selected for external validation, only VEGFR2 (rs1870377) was significantly associated with outcome in our dataset. VEGFR2 (rs1870377) was associated with PFS (median 2.9 months AA, 5.3 months AT, 5.3 months TT, P=0.048, HR 1.35, 95% CI: 1.006–1.822) but not OS. In the paper by Zheng et al. (25) from which the SNP was selected, VEGFR2 (rs1870377) AA was prognostic for TTP and OS but with a HR less than 1 suggesting a protective effect which was opposite in direction to the effect found in our dataset raising doubts about the validity of the prognostic value of the variant. rs1870377 T->A results in a missense mutation resulting in an amino acid alteration in the extracellular domain of VEGFR-2, 472H>Q which has been shown to decrease the affinity of VEGFR-2 for its ligand VEGF (26). In a study by Zhu et al. (27) VEGFR-2 SNPs were studied as prognostic biomarkers in patients with resected gastric cancer and found rs1870377 was prognostic (AA vs. AT, HR 1.69). Zhuo et al. (28) found that in advanced gastric cancer patients rs1870377 was associated with OS (AA/AT vs. TT HR 2.04). Interestingly, there have been other studies showing a prognostic implication of rs1870377 in patients with advanced cancer receiving treatment with anti-VEGF therapies. Maeng et al. (29) found that in patients with advanced gastric or biliary cancers receiving sunitinib, rs1870377 was associated with prognosis AA vs. AT/TT HR 2.27). Gal et al. (30) studied patients with metastatic breast cancer treated with fist line paclitaxel and bevacizumab and round rs1870377 was prognostic for OS (AA/AT vs. TT HR 1.69). These results are congruent with the findings of our study suggesting this SNP may be a relevant biomarker in patients undergoing anti-VEGF therapies.
Significant basic science and clinical research has suggested an important role of the tumor immune microenvironment, and its crosstalk with angiogenesis in the prognosis of HCC. Therefore, an exploratory analysis of immunomodulatory SNPs was carried out to determine their prognostic value in patients with advanced HCC treated with sorafenib (Table S2). SNPs in four immunomodulatory genes were found to be prognostic in our dataset, including CCL2, IL-10, CTLA-4 and NFKB1.
An association was found for the CCL2 SNP rs1024611 for OS (HR 0.67, G vs. A) with no association for PFS. Functionally, rs1024611 is located in the 5' UTR of CCL2. rs1024611 influences the expression of CCL2 through allelic expression imbalance with preferential expression of the G allele. An in vitro study found A>G lead to lower levels of CCL2 in a dose dependent manner (31). Tse et al. (32) found an association between the rs1024611 GG genotype and CCL2 expression in nasopharyngeal tumors and with tumor macrophage infiltration. rs1024611 was also associated with the development of distant metastasis in patients treated with radiotherapy. Patients with the AA or AG genotype had a worse distant metastasis free survival (DMFS) compared to the GG genotype (HR 2.21, or 2.23 respectively).
An association was found with the CTLA-4 SNP rs231775 and PFS (HR 1.17 G vs. A) but not with OS. Functionally, rs231775 results in a missense mutation in CTLA-4, with the G allele leading to decreased expression at the cell surface (33). Liu et al. (33) studied rs231775 in patients with advanced RCC patients receiving sunitinib and found an association with OS (GG vs. AG/AA HR 0.83) however directionally the effect was opposite to that seen in our analysis, with patients with the GG genotype associated with improved prognosis. Similarly, Machado-Rugolo et al. (34) found an association of rs231775 with prognosis in patients with advanced NSCLC but with an opposite direction of effect as found in our analysis.
An association was found for the IL-10 rs1800896 for both OS (HR 0.87 C vs. T) and PFS (HR 0.77 C vs. T). Functionally, rs1800896 has two potential functional effects. On the plus strand, rs1800896 is upstream of IL-10, with the G allele being associated with increased levels of IL-10 mRNA (35). On the minus strand, rs1800896 is an intronic variant within IL-19. rs1800896 does not have well documented prognostic implications in other cancer types in the literature (36).
An association was found with the NFKB1 SNP, rs28362491 and OS (HR 1.14 DEL vs. ATTG) as well as with PFS (HR 1.35 DEL vs. ATTG). Functionally encodes for NFKB1 insertion/deletion in the promoter region, and prior work has shown the deletion variant to be associated with less promoter activity (37). The literature has not described an association between this polymorphism and prognosis in other cancer types.
SNPs were tested for prognostic value in patients with advanced HCC treated with TACE. Of the SNPs found to be prognostic in sorafenib treated patients, only NFKB1 rs28362491 was also found to be prognostic for TACE treated patients but the HR directions was opposite to that observed in the sorafenib treated patients. This finding can be considered hypothesis generating that the identified SNPs in the sorafenib treated patients may be predictive to sorafenib effect, however this would require confirmation, ideally in a trial in which patients were randomized to either receive or not receive sorafenib.
Implications and actions needed
Although our dataset did not validate any of the selected SNPs, VEGFR2 (rs1870377) deserves further study in an independent dataset as a prognostic factor because our results corroborate the effect seen in studies other tumor sites. Furthermore, CCL2 (rs1024611), IL-10 (rs1800896), CTLA-4 (rs231775) and NFKB1 (rs28362491) should be validated in external datasets and in patients treated with modern therapies. If confirmed, these SNPs could be retrospectively evaluated in randomized trials of sorafenib to determine if they are only prognostic or may be predictive of response to sorafenib. If validated these SNPs could also be incorporated into existing prognostic scoring systems and inform basic science investigations to better understand the HCC TME.
Conclusions
In summary, HCC is a global, highly prevalent disease with poor treatment outcomes and limited biomarkers available to guide clinicians on prognosis. We failed to validate prognostic SNPs from the literature in our dataset. We did find that one SNP, VEGFR2 (rs1870377) was significantly associated with outcome in our dataset, however the direction of effect was opposite to that in the source paper. Further literature review revealed that VEGFR2 (rs1870377) is prognostic in other cancer types treated with anti-VEGF therapies with a similar direction of effect as in our data. These findings underscore the difficulty repeating SNP results in observational studies given the potential for spurious associations when multiple variants are tested, effect sizes are not large or when sample sizes are low. We identified SNPs with functional impact on immune signaling in the HCC TME and tested their prognostic significance in sorafenib treated patients. We found that CCL2 (rs1024611), IL-10 (rs1800896), CTLA-4 (rs231775) and NFKB1 (rs28362491) were prognostic in our dataset. Finally, each of the validation and exploratory SNPs were tested in TACE treated patients and were not found to be prognostic, indicating a possible interaction effect with treatment modality.
Acknowledgments
We thank Wei Xu, Biostatistics Department, Dhalla Lana School of Public Health, University of Toronto who assisted with statistical design, discussion of building models and testing data assumptions of this study.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-22/rc
Data Sharing Statement: Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-22/dss
Peer Review File: Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-22/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-22/coif). BHL reports grants, honoraria, and meeting support from Astrazeneca, and grants from Pfizer. DC reports grants, honoraria, or advisory board support from NCI, CIHR, CCSRI, Astrazeneca, Takeda, Boehringer Ingelheim, AMGEN, EMD Serono, Pfizer, Jazz, Merck, Abvie, Anheart, Roche, BMS, and Novartis. GL reports grants, honoraria, or advisory board support from NCI, CIHR, CCSRI, Astrazeneca, Takeda, Boehringer Ingelheim, AMGEN, EMD Serono, Pfizer, Jazz, Merck, Abvie, Anheart, Roche, BMS, and Novartis. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Princess Margaret Cancer Centre (No. IRB00001258) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021;73:4-13. [Crossref] [PubMed]
- Semela D, Dufour JF. Angiogenesis and hepatocellular carcinoma. J Hepatol 2004;41:864-80. [Crossref] [PubMed]
- Morse MA, Sun W, Kim R, et al. The Role of Angiogenesis in Hepatocellular Carcinoma. Clin Cancer Res 2019;25:912-20. [Crossref] [PubMed]
- Fu Y, Liu S, Zeng S, et al. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J Exp Clin Cancer Res 2019;38:396. [Crossref] [PubMed]
- Voron T, Marcheteau E, Pernot S, et al. Control of the immune response by pro-angiogenic factors. Front Oncol 2014;4:70. [Crossref] [PubMed]
- Almand B, Clark JI, Nikitina E, et al. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol 2001;166:678-89. [Crossref] [PubMed]
- Terme M, Pernot S, Marcheteau E, et al. VEGFA-VEGFR pathway blockade inhibits tumor-induced regulatory T-cell proliferation in colorectal cancer. Cancer Res 2013;73:539-49. [Crossref] [PubMed]
- Riabov V, Gudima A, Wang N, et al. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front Physiol 2014;5:75. [Crossref] [PubMed]
- Nagakawa Y, Aoki T, Kasuya K, et al. Histologic features of venous invasion, expression of vascular endothelial growth factor and matrix metalloproteinase-2 and matrix metalloproteinase-9, and the relation with liver metastasis in pancreatic cancer. Pancreas 2002;24:169-78. [Crossref] [PubMed]
- Murdoch C, Muthana M, Coffelt SB, et al. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer 2008;8:618-31. [Crossref] [PubMed]
- Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med 2020;382:1894-905. [Crossref] [PubMed]
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90. [Crossref] [PubMed]
- Gadaleta-Caldarola G, Divella R, Mazzocca A, et al. Sorafenib: the gold standard therapy in advanced hepatocellular carcinoma and beyond. Future Oncol 2015;11:2263-6. [Crossref] [PubMed]
- Tandon P, Garcia-Tsao G. Prognostic indicators in hepatocellular carcinoma: a systematic review of 72 studies. Liver Int 2009;29:502-10. [Crossref] [PubMed]
- Penha Mesquita A, Victor Oliveira Monteiro A, Luiz Araújo Bentes Leal A, et al. Gene variations related to the hepatocellular carcinoma: Results from a field synopsis and Bayesian revaluation. Gene 2023;869:147392. [Crossref] [PubMed]
- Erichsen HC, Chanock SJ. SNPs in cancer research and treatment. Br J Cancer 2004;90:747-51. [Crossref] [PubMed]
- Altman DG, McShane LM, Sauerbrei W, et al. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med 2012;9:e1001216. [Crossref] [PubMed]
- Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723-50. [Crossref] [PubMed]
- Xia SL, Ying SJ, Lin QR, et al. Association of Ulcerative Colitis with FOXP3 Gene Polymorphisms and Its Colonic Expression in Chinese Patients. Gastroenterol Res Pract 2019;2019:4052168. [Crossref] [PubMed]
- Palmirotta R, Carella C, Silvestris E, et al. SNPs in predicting clinical efficacy and toxicity of chemotherapy: walking through the quicksand. Oncotarget 2018;9:25355-82. [Crossref] [PubMed]
- Van den Broeck T, Joniau S, Clinckemalie L, et al. The role of single nucleotide polymorphisms in predicting prostate cancer risk and therapeutic decision making. Biomed Res Int 2014;2014:627510. [Crossref] [PubMed]
- Casadei Gardini A, Marisi G, Faloppi L, et al. eNOS polymorphisms and clinical outcome in advanced HCC patients receiving sorafenib: final results of the ePHAS study. Oncotarget 2016;7:27988-99. [Crossref] [PubMed]
- Marisi G, Petracci E, Raimondi F, et al. ANGPT2 and NOS3 Polymorphisms and Clinical Outcome in Advanced Hepatocellular Carcinoma Patients Receiving Sorafenib. Cancers (Basel) 2019;11:1023. [Crossref] [PubMed]
- Scartozzi M, Faloppi L, Svegliati Baroni G, et al. VEGF and VEGFR genotyping in the prediction of clinical outcome for HCC patients receiving sorafenib: the ALICE-1 study. Int J Cancer 2014;135:1247-56. [Crossref] [PubMed]
- Zheng YB, Zhan MX, Zhao W, et al. The relationship of kinase insert domain receptor gene polymorphisms and clinical outcome in advanced hepatocellular carcinoma patients treated with sorafenib. Med Oncol 2014;31:209. [Crossref] [PubMed]
- Wang Y, Zheng Y, Zhang W, et al. Polymorphisms of KDR gene are associated with coronary heart disease. J Am Coll Cardiol 2007;50:760-7. [Crossref] [PubMed]
- Zhu X, Wang Y, Xue W, et al. The VEGFR-2 protein and the VEGFR-2 rs1870377 A>T genetic polymorphism are prognostic factors for gastric cancer. Cancer Biol Ther 2019;20:497-504. [Crossref] [PubMed]
- Zhuo YJ, Shi Y, Wu T. NRP-1 and KDR polymorphisms are associated with survival time in patients with advanced gastric cancer. Oncol Lett 2019;18:4629-38. [Crossref] [PubMed]
- Maeng CH, Yi JH, Lee J, et al. Effects of single nucleotide polymorphisms on treatment outcomes and toxicity in patients treated with sunitinib. Anticancer Res 2013;33:4619-26. [PubMed]
- Gal J, Milano G, Brest P, et al. VEGF-Related Germinal Polymorphisms May Identify a Subgroup of Breast Cancer Patients with Favorable Outcome under Bevacizumab-Based Therapy-A Message from COMET, a French Unicancer Multicentric Study. Pharmaceuticals (Basel) 2020;13:414. [Crossref] [PubMed]
- Pham MH, Bonello GB, Castiblanco J, et al. The rs1024611 regulatory region polymorphism is associated with CCL2 allelic expression imbalance. PLoS One 2012;7:e49498. [Crossref] [PubMed]
- Tse KP, Tsang NM, Chen KD, et al. MCP-1 Promoter Polymorphism at 2518 is associated with metastasis of nasopharyngeal carcinoma after treatment. Clin Cancer Res 2007;13:6320-6. [Crossref] [PubMed]
- Liu X, Swen JJ, Diekstra MHM, et al. A Genetic Polymorphism in CTLA-4 Is Associated with Overall Survival in Sunitinib-Treated Patients with Clear Cell Metastatic Renal Cell Carcinoma. Clin Cancer Res 2018;24:2350-6. [Crossref] [PubMed]
- Machado-Rugolo J, Fabro A, Cuentas E, et al. P3.04-03 Association of Functional Polymorphism in CTLA-4 Gene with Survival in Non-Small Cell Lung Cancer: A Brazilian Study. J Thorac Oncol 2018;13:S923. [Crossref]
- Iyer SS, Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol 2012;32:23-63. [Crossref] [PubMed]
- Teresa Gonzalez-Garza M, Elva Cruz-Vega D, Maldonado-Bernal C. IL10 as Cancer Biomarker. In: Translational Research in Cancer. IntechOpen; 2021.
- Karban AS, Okazaki T, Panhuysen CI, et al. Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Hum Mol Genet 2004;13:35-45. [Crossref] [PubMed]
Cite this article as: Herman M, Lok BH, Gallinger S, Dawson L, Kim R, Cheng D, Paton T, Bucur R, Patel D, Fazelzad R, Hueniken K, Liu G. Analysis of prognostic germline polymorphisms in patients with advanced hepatocellular carcinoma. Transl Gastroenterol Hepatol 2023;8:32.


