
Novel approaches to liver disease diagnosis and modeling
A multi-omic view of the liver
Liver physiology can be understood as a continuum involving a plethora of processes and regulatory mechanisms at different scales. In this scenario, genes are at the lowest scale and the organ as a whole at the highest scale. The intermediate scales are occupied by RNAs, proteins, metabolites, biological networks, cellular organelles and different cell types. The interplay between the organization scales occurs through multidirectional relations of causation (1,2), whereas, the environment influences processes at lower scales, including the activation of protein networks and gene expression (Figure 1). Ultimately, liver physiology is the result of the tight balance of this complex system characterized by the multiplicity of interactions. Thus, perturbations in any level of organization or in their interaction, usually lead to liver diseases.
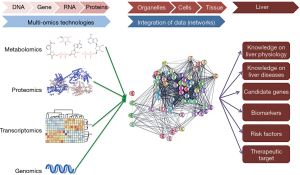
The traditional way by which scientists investigate either physiology or diseases of the liver involves the simplification of this biological system and its byproducts to a single or small group of the constituents at each scale. Although the classical methods allow logical conclusions regarding the role of specific target(s) of interest in any given process, they are not considering the hepatic system as a whole and, therefore, have limited clinical outcomes.
More recently however, hepatology research has shifted from this reductionist view towards more holistic and integrated approaches. The development of new omics technologies (i.e., genomic, proteomic, metabolomic) has allowed scientists to collect more information regarding molecular events in biological systems by addressing large ensembles of components such as genes, proteins, and metabolites. Moreover, these technologies are in constant development and optimization, resulting in increased data output per run at lower cost, which make them far more accessible for researchers, clinicians and their patients (3-5). Finally, the introduction of new bioinformatics tools has increased our ability to integrate information obtained by different omics and empower the application of systems biology in research and clinical practice. Indeed, as will be discussed below, it is the combination of these different technologies that will contribute significantly to the understanding, diagnosis and treatment of liver diseases.
Genomics
A great advance in the hepatology field was the use of genome wide association studies (GWAS), also known as whole genome association studies, that focus on identifying genetic variants predisposing a patient to certain diseases. GWAS has been used in different study designs (family-based, population-based or case-control studies) to screen the frequency of more than 1 million known single nucleotide polymorphisms (SNPs) and have resulted in the discovery of risk loci associated with common and rare liver diseases such as gallstone disease, non-alcoholic fatty liver disease (NAFLD), drug-induced liver injury (DILI), chronic hepatitis C after treatment, cystic fibrosis associated liver disease, Alagille syndrome, Biliary Atresia, auto-imune hepatitis (AIH), primary biliary cholangitis (PBC) and PSC primary sclerosing cholangitis (6-8). Up to date of this manuscript preparation, the NHGRI-EBI catalog of published GWAS studies (www.genome.gov/gwastudies/) reports, under the search term “liver disease”, a total of about 4,493 studies, 120,219 SNPs and 179,364 associations identified. GWAS were shown to have an accuracy ranging between 21% and 51% in the identification of disease-causing SNPs depending on the disease, study design and availability of premade panels of SNPs (9). An important limitation of GWAS studies is that, in many cases, the identified loci might contain several genes that equally contribute to but not necessarily cause the disease. Overall, GWAS studies have been instrumental as a starting point to identify biological pathways associated with multifactorial diseases to be further investigated, but in large part have hampered our expectation to apply this information into clinical practice. This particularly relates to GWAS studies focused on complex diseases. Examples in the liver context are PBC (115 associations and 8 studies) and PSC (308 associations and 17 studies), in which most of the identified disease variants appeared at a very low effect size, suggesting a marginal genetic contribution in respect to other risk factors (i.e., environmental) at play (10,11).
A possible outcome of GWAS has been the establishment of targeted gene panels associated with a certain liver disease. These panels can improve the coverage of the genes of interest and be cost and time effective, finding their application in diagnostic processes including next generation sequencing (NGS) (12). Targeted gene sequencing (TGS) allows early diagnosis and the identification of the specific genetic causes of a disease even within a group of disorders. This approach has been advantageous in pediatric hepatology to identify specific subtypes of cholestatic disorders. Pediatric cholestasis includes a group of disorders with different etiologies (genetic and metabolic) that result in the impairment of bile acid transport and lead to liver failure. Among the inherited conditions, for example, patients with progressive familial intrahepatic cholestasis (PFIC) caused by different genes, present low serum concentrations of gamma-glutamyl transpeptidase (GGT) and are clinically undistinguishable. However, each specific PFIC subtype is associated with different risk factors that determine their clinical management and post-transplant complications (12). In a recent study Nicastro et al., showed the use of a specific NGS panel for genetic liver diseases in pediatric cholestasis, as a time and cost effective diagnostic tool compared to traditional diagnostic, in a transplant center in Italy (13).
Another important genomic tool is whole exome sequencing (WES), an exploratory technique that consists in the sequencing of the protein coding regions of genes—known as exome. The exome consists of approximately 1% of the human genome but harbors 85% of the mutations with effects on disease traits (14). The cost of sequencing in WES is lower than in whole genome sequencing (WGS), making the former technology a promising tool for early diagnosis in clinic. A recent study in a small number of patients showed the applicability and cost-effectiveness of WES in clinical practice to identify disease-related variants in adults with unexplained liver disease (15). Hakim et al. applied WES for the diagnosis and the identification of molecular causes in 19 adult patients with chronic liver disease of unknown etiology. The genomic analysis brought to the identification of the disease diagnosis and predict the correct treatment in 5 of these cases (15).
Although the use of genomic techniques, such as TGS, WES and WGS awaits further validation for larger population studies, it certainly shows promise as a useful tool for early and precise diagnosis and better clinical management of genetic liver diseases. This highlights the importance of genome sequencing in research and clinical practice and how NGS technologies can revolutionize medicine. On the other hand, studies that carefully analyze the real cost effectiveness of NGS technologies are necessary before they can be fully integrated into clinical practice (16). Moreover, the applicability of these technologies remains limited in the diagnosis of pathologies in which the genetic factor is less prominent and even in the determination of which identified variant(s) contribute(s) more to the trait or disease of interest (6). To overcome these limitations, GWAS can be associated with transcriptomics to inform on risk variants, gene expression profiling and altered pathways associated with the disease (17-19).
The transcriptome can be defined as the complete set, and quantity, of transcripts within a cell, tissue or organ in physiological or pathological conditions (20) and transcriptomics are the set of technologies used to profile the transcriptome. Since its development, RNA sequencing (RNAseq) technology has become the most widely used technology to investigate the transcriptome of a target sample. In general terms, a cDNA library is assembled from the extracted RNA and then sequenced starting from one end (single ended sequencing) or both ends (paired ended sequencing). The result is a high-throughput dataset of sequences (reads), whose number and length depend on the sequencing strategy used. Then, reads are processed to remove poor quality regions, sequencing adapters or any other technical sequences to optimize downstream analysis (21). Following this step, the trimmed reads are aligned against a reference (indexed) genome, which allows the accurate counting of expressed genes (22). Importantly, there are many bioinformatics tools freely available to the scientific community that can be used in each of these steps; as well as up-to-date and curated sequence and annotation for the genomes of human and common experimental animal models such as mouse (Mus musculus) and rat (Rattus norvegicus). Finally, the investigator will perform a series of statistical analysis to detect differentially expressed genes among groups and the overrepresented terms or signaling pathways associated with those genes. Ultimately, the result is a picture of the expression landscape between 2 or more conditions that can provide important information on the role of individual genes or gene clusters to the phenotype of interest (20-22).
An advance in the analysis of transcriptome is the single cell RNAseq (scRNAseq). As the name implies, scRNAseq increases the resolution of information, as now it is possible to perform differential expression analysis in individual cells isolated from an organ such as the liver; as well as clustering, classification and dynamics of different cell types in different conditions or timepoints (23,24). By using this approach, researchers are now providing detailed maps of the cellular transcriptional landscape and heterogeneity in the liver during development and in adulthood under physiological conditions (25,26) or during different liver diseases, as exemplified by hepatocellular carcinoma, non-alcoholic steatohepatitis (NASH) and fibrosis (27-29). These approaches will surely contribute to the understanding of liver physiology and pathologies, in particular for rare diseases, where the limited number of patients prevents the use of other genomic tools. In line with this, transcriptomics can be used to unravel the mechanisms of current treatments (30), or as a tool for drug repositioning (31) or identification of new therapeutic targets (32,33).
Proteomics
Ideally the data of gene variants or altered gene expression obtained from genomics or transcriptomics analysis should be correlated to differences in protein abundance. Indeed, a recent example of this correlation has been demonstrated for the mannose phosphate isomerase (MPI) gene in a pediatric population. Transcriptomic data from three human liver cohorts showed that in children with mutations in the MPI gene, a rare monogenic disorder, the MPI expression level is proportional to liver fibrosis (30).
Although gene-protein correlations are useful in many instances, they are by no means linear. Posttranscriptional and posttranslational mechanisms alter this correlation by influencing the abundance, function, and networks of proteins. It is known that these mechanisms contribute to the greater complexity and larger diversity of proteins when compared to genes (34). Therefore, a more comprehensive approach to studying the whole set of proteins within a biological system, or proteome, is through its large-scale analysis, or proteomics. Currently, mass spectrometry (MS)-based proteomics is the most commonly used technology to address the proteome (35). This technique comprises the isolation of proteins from the sample, its downstream processing and the analysis in a high-resolution mass spectrometer. The result is a mass-charge (m/z) spectrum that is used in protein identification and quantification. There are different strategies for proteomics (bottom-up and top-down proteomics), which result in distinct outcomes as global protein abundance and/or posttranslational modifications, including phosphorylation, glycosylation, and others. In this exploratory approach, in which the whole ensemble of proteins is the background, the investigator may submit the results to downstream analysis to detect differentially abundant proteins, interaction networks, up or downstream signaling pathways, enriched terms or pathways and visualization of the proteins in the context of overrepresented biologically-significant terms. Thus, the analyses will depend on the biological question to be addressed.
The application of proteomics in hepatology thus far has been limited to physiological studies. For example, a detailed characterization of the proteome and protein networks from different hepatic cell types was performed and revealed the complementarity and the crosstalk of these networks among different cell types (36), which is a noteworthy feature of liver organization. Using a similar approach, another study established that a large proportion of the hepatocyte proteome is involved in fatty acid and xenobiotic metabolism. It also revealed that different populations of hepatocytes play distinct roles in cholesterol flux and hormone/growth factors signaling (37).
In the context of liver diseases, MS-based proteomics is an important tool for the identification of putative pathogenic proteins. Analyses of serum from patients with autoimmune hepatitis (AIH) identified antigenic proteins associated with the disease, including phosphoglycerate mutase isozyme B, liver arginase, cytokeratins 8 and 18, heat shock proteins and valosin-containing protein (38). Furthermore, proteomics revealed overrepresented targets related to Th1 and Th2 type responses through analysis of the immune response signatures in this disease (39). These key findings identified new candidate antigens, molecular pathways, biomarkers and even diagnostic models with high sensitivity and specificity for liver autoimmune diseases (40-42).
Proteomics can also be useful to identify biomarkers for the diagnosis and the progression of a specific diseases. For example, saliva proteome analysis using an integrated top-down/bottom-up platform was used in a population of patients with Wilson’s disease, a rare inherited disorder of copper metabolism that include liver symptoms (43). This analysis showed that the proteins present in the saliva of these patients are representative of the oxidative stress and inflammatory condition typical of the disease and could potentially be a marker of exacerbation of the disease (43).
In another interesting study, Metzger et al. used urine proteomics to differentiate cholangiocarcinoma (CC) from PSC and other benign biliary diseases (BBD) (44). Knowing from a previous study that bile proteomic can distinguish CC from PSC, the urine was compared with that of the bile of the same patients collected in the same day. The study identified urine proteins that are able to distinguish CC from PSC and BBD suggesting that urine proteomic analysis (compared to bile) can be used as a non-invasive diagnostic tool for PSC patients at risk of developing CC (44).
A related technique known as targeted proteomics involves the investigation of a specific set of proteins of interest known to be associated with certain phenotypes or diseases. In this approach, the background is composed of sets of hundreds of proteins rather than the thousands that comprise the whole proteome. Because of its lower complexity (vs. “non-targeted” proteomics), as well as better scalability, sensitivity, precision, accuracy and reproducibility (45), targeted-proteomics can be useful not only in research, but also in precision and personalized medicine (46). It can be used, for example, in the diagnosis, as well as in the clinical management of complex or rare liver diseases.
Metabolomics
Although powerful, transcriptomics and proteomics are only a surrogate picture of the physiological outcomes of a certain cluster of differentially expressed targets or overrepresented pathways. A complete and more robust physiological snapshot can be achieved by the application of metabolomics: the high-throughput identification and analysis of the set of small molecule metabolites (<1,500 Da), known as the metabolome. As a result of metabolomics studies, the metabolic signature has been associated with not only common liver diseases such as NAFLD and liver toxicity (47-51) but also with liver regeneration (52) and in the identification of novel biomarkers (53). As proteomics, metabolomics has also contributed to the identification of novel biomarkers in autoimmune liver diseases and improvements in diagnosis. The main results of these studies showed that metabolites such as glycochenodeoxycholic acid and the free-fatty acids (FFA): LPC-16:0, PC-16:0/16:0 and SM could be used to diagnose autoimmune liver diseases (i.e., AIH and PBC) and PBC, whereas the bile salts LCA-S, TDCA, and GDCA could be used in the differential diagnose of PBC and primary sclerosing cholangitis (PSC), a rare chronic inflammatory cholangiopathy of unknown etiology (54,55). Another study identified citrate, glutamine, acetone, pyruvate, β-hydroxyisobutyrate, acetoacetate, histidine, dimethylamine, and creatinine as metabolites with a high diagnostic accuracy for the discrimination of AIH and PBC (56). Although promising, the results are still preliminary because of the limited number of studies and differences in methodology (57).
Because of the recent advances in the technologies used for non-targeted or targeted identification of metabolites, alongside the development of novel computational analysis protocols, the metabolic phenotyping is now a promising tool in the understanding of liver diseases, in their diagnosis and clinical management, including disease staging and individual monitoring of patient response to available treatments.
Microbiome
Thus far, we have discussed the application of high-throughput-omics approaches in the study of molecular events at different scales of organization focusing in the liver, from genes (lowest scale) to RNA, proteins, and metabolites (intermediate scales) towards a phenotype (Figure 1). Another scaling-up level is to consider exchangeable metabolites and factors produced outside of the liver that can impact its functioning. In line with this interpretation, an important source of exchangeable metabolites to the liver is the large population of microorganisms, the microbiota, that reside in the gastrointestinal tract. This community has its own genome, proteome and metabolites (collectively known as microbiome) that can be greater in extent and diversity than the host’s (58,59). Products from the microbiota can be absorbed into the bloodstream and reach the hepatic tissue through the portal circulation or simply stimulate neuroendocrine signaling pathways to the liver. Therefore, the microbiome may have direct or indirect impact on liver physiology. A correlate term with -omics is the meta-omics (metagenomics, metaproteomics and so on), which refers to the large-scale studies of the microbiome (2). Microbiota and microbiome studies identified a high producing alcohol strain of Klebsiella pneumonia in 60% of patients with NAFLD in a Chinese cohort. Moreover, this strain was able to induce disease in mice (60). Similarly, alterations in dietary fat content resulted in dysbiosis, with consequent alterations in nutrient-microbiota interaction, a scenario in which the production of a specific short-chain fatty acid was associated with liver steatosis and metabolic syndrome (61). Intestinal dysbiosis characterized by a reduced bacterial diversity was also described in patients with PSC (62-64). Recent studies have shown that the dysbiosis is not limited to the fecal and colonic mucosa, but it is also extended to the bile of these patients, showing an increased presence of the phylum Proteobacteria (65). Interestingly, microbiota has been suggested to act as the driver of the increased Th17 differentiation often seen in PSC patients, suggesting that the gut microbiome could be the trigger of adaptive immune reactions (immunobiome) (66).
Panomics
In face of the great amount of data that we now can collect from an organism and the availability of public repositories for such data, a striking challenge is the integration of all this information. The Human Cell Atlas (https://www.humancellatlas.org/), human microbiome project (https://hmpdacc.org/hmp/) and human metabolome database (http://www.hmdb.ca/) are initiatives that exemplify the effort of the scientific community in integrating multi-omics data and improving accessibility to the cumulative knowledge (67,68). The ultimate goal is, with help from machine learning and other computational tools, to integrate high-throughput datasets with other relevant clinical information, including anamnesis and routine biochemical laboratory tests into an algorithm that can accurately predict disease progress, patient response to treatments (69), and prognosis (70).
Disease modeling
The integration of multi-omics data allows the identification of specific molecular targets associated with a disease. However, these targets should ideally be functionally validated using reliable models of the cell or tissue affected by the disease, to specifically address the disease mechanism and potential therapies.
Our current knowledge of altered mechanisms in liver diseases mainly derives from studies using in vivo animal models, in vitro primary cell culture systems and cancer-derived or immortalized cell lines. However, these approaches have several limitations. Studies in rodents suffer very often from a lack of phenotype reproducibility due to inter-species differences (71), whereas the use of in vitro human primary cultures is hampered by the scarce availability of human liver tissues, and in particular for rare liver diseases. An additional limitation is the technical difficulties in handling primary cells cultures. Conversely, immortalized cell lines do not always recapitulate normal physiological functions of the tissue that they are isolated from (72,73). These collective shortcomings of traditional tools to model liver diseases in culture are being addressed by recent advances in stem cell-based in vitro models (i.e., iPSC and 3D organoid cultures) (Figure 2) (74).
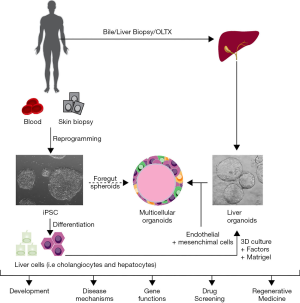
Induced pluripotent stem cells (iPSCs) and applications
The discovery of iPSCs dates back to 2006 when Takahashi and Yamanaka were able to induce a state of pluripotency in skin fibroblasts by exogenous expression of four pluripotency transcription factors (i.e., Klf6, Oct4, Sox2 and c-Myc) (75). The resulting iPSCs were characterized by indefinite self-renewal and by the ability to differentiate into a variety of cell types. More recently, iPSCs have been obtained from other sources such as peripheral blood cells and urine, further decreasing the invasiveness of the procedure (76,77).
In the context of modern medicine, iPSCs find many applications: they can be derived from a patient and used to study the mechanism of a known disease to find specific targets for therapy; or to identify the function of genes associated with a novel disease; and finally to screen compounds in a large scale and to derive specific treatments. This approach would particularly benefit those rare diseases for which clinical trials could not be designed due to a very limited number of patients (78).
In liver research specifically, iPSCs have been mainly used for the generation of hepatocytes and cholangiocytes. Most of the protocols for the differentiation of iPSC into liver cells recapitulate the different steps of liver embryogenesis, characterized by the sequential progression of the pluripotent stem cells into a stage of definitive endoderm, hepatic endoderm and ultimately the hepatoblast, the common bipotent progenitor for hepatocytes and cholangiocytes. This process is mediated by the addition in the culture media of specific factors aimed at activating the different molecular pathways that take place during liver development (79).
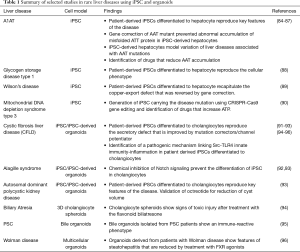
Hepatocytes have been the first liver cells to be generated from iPSC (80-82). However, despite many efforts, as of today, iPSC-derived hepatocyte-like cells lack full maturation and present a more fetal phenotype (83) in comparison to primary hepatocytes. Despite this limitation, many researchers have been able to use iPSC-derived hepatocytes to reproduce and model in vitro liver diseases (Table 1) such as A1AT deficiency (84-87), familial hypercholesterolemia (FH) (97), Glycogen Storage Disease Type 1 (88), Wilson’s disease (89) and mitochondrial DNA depletion syndrome type 3 (90), suggesting that the less mature phenotype is not affecting the disease mechanism itself.
Full table
More recent studies demonstrated that iPSC can also be made to differentiate into biliary cell (91-93,98,99). Compared to hepatocytes, the biliary lineage appears to constitute a more default pathway of differentiation as iPSC-derived biliary cells more closely resemble adult cholangiocytes with preserved secretory functions. iPSC-derived cholangiocytes have been used to model rare diseases of the biliary epithelium or cholangiopathies such as cystic fibrosis related liver disease (CFLD), Alagille syndrome and autosomal dominant polycystic liver disease (91-93) (Table 1).
iPSCs provide a virtually unlimited source of disease cells and therefore they have been used for drug discovery or repurposing to target specific pathways or, on a larger scale, for the discovery of new compounds in a platform of high throughput screening. Using iPSC-derived hepatocytes, 5 new drugs from a library of clinical compounds have been identified for A1AT deficiency (87). In a similar way, cardiac glycosides, currently approved for heart failure, were shown to be a potential candidate to treat hypercholesterolemia, at least in those patients that develop resistance to statins (100).
Another study, using iPSC-derived cholangiocytes from a CF patient carrying the common CFTR mutation DF508, identified a pathogenic mechanism that links a constitutive activation of Src kinase in CFTR defective cholangiocytes, with an aberrant activation of TLR-mediated innate immunity pathways. Interestingly, the study also showed that inhibition of the kinase could increase the efficacy of currently approved potentiator/correctors used in CF patients with this mutation (91).
iPSC can also be genetically modified using CRISPR-Cas9 technology. Using this approach, it is possible to introduce a specific disease-causing mutation in control iPSC or to correct specific mutations in patient-derived iPSC, thus providing isogenic matching controls to characterize the disease phenotype related to a specific mutation (101). This tool is especially important for rare diseases that affect only a few individuals or a monogenic disease with several rare variants (i.e., CF) since it will accelerate their molecular characterization and will create valid platforms for cost effective preclinical drug testing.
Liver organoids and applications
Another important advance of stem cell technology has been the establishment of 3D organotypic culture systems that resemble the architecture and the function of a specific tissue. Compared to traditional 2D cultures (i.e., monolayers), organoids are more physiologically relevant. Organoids have the ability to self-organize in structures that resembles the organ of origin and therefore they reproduce the in vivo cell-cell and cell-matrix interactions. This allows them to better preserve key functional features than in traditional 2D culture. It is well known that primary cells drastically decrease their proliferative capability when in 2D culture while they rapidly de-differentiate, therefore precluding their propagation for multiple passages. Conversely, organoids have shown to be self-renewing and therefore can be propagated for long-term in culture without loss of functionality while maintaining their genetic stability for several passages (74,102,103).
Liver organoids can be derived from different sources of stem cells, including iPSC (104), adult or embryonic tissue-resident stem cells and more recently (105), from bile fluid collected during ERCP (endoscopic retrograde cholangiopancreatography) procedures (95). In order to self-organize in 3D structures, stem cells from any origin need to be embedded in an extracellular matrix (i.e., Matrigel) and be supplied with specific media that contain factors able to recapitulate the in vivo environment of the stem cell niche both in normal conditions (i.e., self-renewal) or during repair/regeneration (103).
To date, liver organoids derived from tissue resident stem cells or from bile samples are more committed to a default biliary phenotype, but more recently mature hepatocyte organoids have been successfully derived from adult primary hepatocytes (106).
Liver organoids, in particular those obtained from patient-derived iPSC, have been used to model specific functional defects representative of monogenic rare liver diseases such as Alagille syndrome, polycystic liver disease, cystic fibrosis, Wilson disease and AAT1 (92,93,105,107) (Table 1) in an attempt to generate specific platforms for drug screening.
More recently, liver organoids have been applied to the study of complex or acquired liver diseases. Using mouse 3D cholangiocyte spheroids exposed to the flavonoid toxin biliatresone, Lorent et al. modeled Biliary Atresia. The treatment with the toxin caused loss of the primary cilia and disruption of the biliary cell polarity, providing evidence of a toxic environmental injury as a potential initiator of the disease (94). In another study, bile organoids have been derived from PSC patients that undergo ERCP as part of their diagnostic and clinical management (95). Further characterization of these organoids showed that they retain key characteristics associated with the disease, such as an immune-reactive phenotype and the ability to secrete inflammatory mediators in response to inflammatory stimuli (95). Finally, liver organoids have also been used to mimic virus-host interaction and study the process of HCV entry into polarized hepatocytes and to create an in vitro HBV infection model (108,109).
It is tempting to speculate that liver organoids will have a major impact on personalized medicine because they can be cryopreserved and bio-banked for later use in drug screening and possibly for regenerative medicine purposes, such as autologous cell therapy transplantation (102,103). Indeed, Schneeberger et al. described in a recent study, a new method based on spinner flasks for the expansion of a large number of liver organoids (110). Using this system, the organoids were able to proliferate at a high rate and to repopulate and reconstitute the liver tissue in decellularized liver discs. The possibility to scale-up the production of liver organoids, together with the application of CRISPR/Cas9 gene editing for the correction of specific mutations will pave the way for the use of regenerative medicine in the treatment of rare diseases.
As promising as stem cell technologies are, some limitations still remain. One is the homogeneous cell composition of liver organoids that do not completely recapitulate the multiple cell types of the liver. To that end, Takebe’s group has made important steps toward the realization of more complex systems. In an initial study, they showed the association of iPSC-derived hepatoblasts with human umbilical endothelial cells (HUVECs) and human bone marrow-derived mesenchymal cells that generated foetal liver buds, which are able to engraft and vascularize in NOD/SCID mice (111). In a follow-up study, they showed the capability of liver bud formation by combining endodermal, endothelial and mesenchymal cells entirely derived from iPSC (112). In a more recent study, they were able to generate multi-cellular human liver organoids (HLOs) starting from foregut spheroids in a single protocol. The generated organoids contained epithelial cells (i.e., expressing hepatocyte and cholangiocyte markers) but also cells of mesenchymal origin (i.e., expressing stellate and Kupffer cell markers). Interestingly, in organoids derived from iPSC of patients with Wolman disease, a rare genetic condition characterized by a defective activity of lysosomal acid lipase, they found features of steatohepatitis that could be rescued by treatment with FXR agonists (96). A limitation of the study is that the functional activity of each cell type was not fully characterized. Finally, the same group modeled a multi-organ (hepato-biliary-pancreatic) integrated system starting from iPSC spheroids, for the study of human organogenesis (113). Overall, these models are ways from having a clinical application and many challenges need to be overcome. However, they demonstrate how stem cell technology is fast evolving and how it might contribute to translational medicine.
Another limitation not yet completely solved is the need of ECMs, whose composition is not well defined, to initiate the 3D aggregation. In that regard, bioengineers are making a remarkable effort to develop new hydrogels with defined composition that support organoids formation and growth (114).
Concluding remarks
With the rapid evolution of omics technologies, the acquisition of new tools for data analysis and integration and the continued reduction of costs for such a large-scale approach, we will witness their introduction in the clinical practice in the near future. When combined, multi-omics analysis will provide a useful snapshot of the disease mechanism and eventually be applied for the prediction of disease risk, for early diagnosis and the development of potential therapeutic targets. In this context, stem cell-derived human models (i.e., iPSC and organoids) will help to tailor these approaches into biomedical applications for personalized medicine.
Acknowledgments
Funding: RF was supported by Cystic Fibrosis Foundation [FIOROT8GO] and by the National Institutes of Health (2 RO1DK096096-06A1). AGO was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado de Minas (FAPEMIG), Brazil.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Luca Fabris and Mario Strazzabosco) for the series “Recent Advances in Rare Liver Diseases” published in Translational Gastroenterology and Hepatology. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tgh-20-109). The series “Recent Advances in Rare Liver Diseases” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Noble D. A theory of biological relativity: no privileged level of causation. Interface Focus 2012;2:55-64. [Crossref] [PubMed]
- Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome 2015;3:31. [Crossref] [PubMed]
- DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). Available online: www.genome.gov/sequencingcostsdata
- Mardis ER. A decade's perspective on DNA sequencing technology. Nature 2011;470:198-203. [Crossref] [PubMed]
- Weymann D, Laskin J, Roscoe R, et al. The cost and cost trajectory of whole-genome analysis guiding treatment of patients with advanced cancers. Mol Genet Genomic Med 2017;5:251-60. [Crossref] [PubMed]
- Hirschfield GM, Chapman RW, Karlsen TH, et al. The genetics of complex cholestatic disorders. Gastroenterology 2013;144:1357-74. [Crossref] [PubMed]
- Karlsen TH, Lammert F, Thompson RJ. Genetics of liver disease: From pathophysiology to clinical practice. J Hepatol 2015;62:S6-S14. [Crossref] [PubMed]
- Krawczyk M, Müllenbach R, Weber SN, et al. Genome-wide association studies and genetic risk assessment of liver diseases. Nat Rev Gastroenterol Hepatol 2010;7:669-81. [Crossref] [PubMed]
- Taylor JC, Martin HC, Lise S, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet 2015;47:717-26. [Crossref] [PubMed]
- Chung BK, Karlsen TH. Genetic Discoveries Highlight Environmental Factors as Key Drivers of Liver Disease. Dig Dis 2017;35:323-33. [Crossref] [PubMed]
- Karlsen TH, Chung BK. Genetic Risk and the Development of Autoimmune Liver Disease. Dig Dis 2015;33 Suppl 2:13-24. [Crossref] [PubMed]
- Nicastro E, D'Antiga L. Next generation sequencing in pediatric hepatology and liver transplantation. Liver Transpl 2018;24:282-93. [Crossref] [PubMed]
- Nicastro E, Di Giorgio A, Marchetti D, et al. Diagnostic Yield of an Algorithm for Neonatal and Infantile Cholestasis Integrating Next-Generation Sequencing. J Pediatr 2019;211:54-62.e4. [Crossref] [PubMed]
- Lalonde E, Albrecht S, Ha KC, et al. Unexpected allelic heterogeneity and spectrum of mutations in Fowler syndrome revealed by next-generation exome sequencing. Hum Mutat 2010;31:918-23. [Crossref] [PubMed]
- Hakim A, Zhang X, DeLisle A, et al. Clinical utility of genomic analysis in adults with idiopathic liver disease. J Hepatol 2019;70:1214-21. [Crossref] [PubMed]
- Schwarze K, Buchanan J, Taylor JC, et al. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med 2018;20:1122-30. [Crossref] [PubMed]
- Caliskan M, Manduchi E, Rao HS, et al. Genetic and Epigenetic Fine Mapping of Complex Trait Associated Loci in the Human Liver. Am J Hum Genet 2019;105:89-107. [Crossref] [PubMed]
- Liu ZH, Lian BF, Dong QZ, et al. Whole-exome mutational and transcriptional landscapes of combined hepatocellular cholangiocarcinoma and intrahepatic cholangiocarcinoma reveal molecular diversity. Biochim Biophys Acta Mol Basis Dis 2018;1864:2360-8. [Crossref] [PubMed]
- Smalling RL, Delker DA, Zhang Y, et al. Genome-wide transcriptome analysis identifies novel gene signatures implicated in human chronic liver disease. Am J Physiol Gastrointest Liver Physiol 2013;305:G364-74. [Crossref] [PubMed]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57-63. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15-21. [Crossref] [PubMed]
- Vieth B, Parekh S, Ziegenhain C, et al. A systematic evaluation of single cell RNA-seq analysis pipelines. Nat Commun 2019;10:4667. [Crossref] [PubMed]
- Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol 2016;34:1145-60. [Crossref] [PubMed]
- MacParland SA, Liu JC, Ma XZ, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 2018;9:4383. [Crossref] [PubMed]
- Su X, Shi Y, Zou X, et al. Single-cell RNA-Seq analysis reveals dynamic trajectories during mouse liver development. BMC Genomics 2017;18:946. [Crossref] [PubMed]
- Ramachandran P, Dobie R, Wilson-Kanamori JR, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019;575:512-8. [Crossref] [PubMed]
- Xiong X, Kuang H, Ansari S, et al. Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol Cell 2019;75:644-60.e5. [Crossref] [PubMed]
- Zheng C, Zheng L, Yoo JK, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017;169:1342-56.e16. [Crossref] [PubMed]
- DeRossi C, Bambino K, Morrison J, et al. Mannose Phosphate Isomerase and Mannose Regulate Hepatic Stellate Cell Activation and Fibrosis in Zebrafish and Humans. Hepatology 2019;70:2107-22. [Crossref] [PubMed]
- Chen B, Wei W, Ma L, et al. Computational Discovery of Niclosamide Ethanolamine, a Repurposed Drug Candidate That Reduces Growth of Hepatocellular Carcinoma Cells In Vitro and in Mice by Inhibiting Cell Division Cycle 37 Signaling. Gastroenterology 2017;152:2022-36. [Crossref] [PubMed]
- Bi X, Pashos EE, Cuchel M, et al. ATP-Binding Cassette Transporter A1 Deficiency in Human Induced Pluripotent Stem Cell-Derived Hepatocytes Abrogates HDL Biogenesis and Enhances Triglyceride Secretion. EBioMedicine 2017;18:139-45. [Crossref] [PubMed]
- Yu FPS, Molino S, Sikora J, et al. Hepatic pathology and altered gene transcription in a murine model of acid ceramidase deficiency. Lab Invest 2019;99:1572-92. [Crossref] [PubMed]
- Zhang Z, Wu S, Stenoien DL, et al. High-throughput proteomics. Annu Rev Anal Chem (Palo Alto Calif) 2014;7:427-54. [Crossref] [PubMed]
- Li X, Wang W, Chen J. Recent progress in mass spectrometry proteomics for biomedical research. Sci China Life Sci 2017;60:1093-113. [Crossref] [PubMed]
- Ding C, Li Y, Guo F, et al. A Cell-type-resolved Liver Proteome. Mol Cell Proteomics 2016;15:3190-202. [Crossref] [PubMed]
- Azimifar SB, Nagaraj N, Cox J, et al. Cell-type-resolved quantitative proteomics of murine liver. Cell Metab 2014;20:1076-87. [Crossref] [PubMed]
- Tahiri F, Le Naour F, Huguet S, et al. Identification of plasma membrane autoantigens in autoimmune hepatitis type 1 using a proteomics tool. Hepatology 2008;47:937-48. [Crossref] [PubMed]
- Shackel NA, McGuinness PH, Abbott CA, et al. Identification of novel molecules and pathogenic pathways in primary biliary cirrhosis: cDNA array analysis of intrahepatic differential gene expression. Gut 2001;49:565-76. [Crossref] [PubMed]
- Czaja AJ. Autoimmune liver disease. Curr Opin Gastroenterol 2009;25:215-22. [Crossref] [PubMed]
- Hannivoort RA, Hernandez-Gea V, Friedman SL. Genomics and proteomics in liver fibrosis and cirrhosis. Fibrogenesis Tissue Repair 2012;5:1. [Crossref] [PubMed]
- Li YZ, Hu CJ, Leng XM, et al. Promising diagnostic biomarkers for primary biliary cirrhosis identified with magnetic beads and MALDI-TOF-MS. Anat Rec (Hoboken) 2009;292:455-60. [Crossref] [PubMed]
- Cabras T, Sanna M, Manconi B, et al. Proteomic investigation of whole saliva in Wilson's disease. J Proteomics 2015;128:154-63. [Crossref] [PubMed]
- Metzger J, Negm AA, Plentz RR, et al. Urine proteomic analysis differentiates cholangiocarcinoma from primary sclerosing cholangitis and other benign biliary disorders. Gut 2013;62:122-30. [Crossref] [PubMed]
- Uzozie AC, Aebersold R. Advancing translational research and precision medicine with targeted proteomics. J Proteomics 2018;189:1-10. [Crossref] [PubMed]
- Forler S, Klein O, Klose J. Individualized proteomics. J Proteomics 2014;107:56-61. [Crossref] [PubMed]
- Deng P, Barney J, Petriello MC, et al. Hepatic metabolomics reveals that liver injury increases PCB 126-induced oxidative stress and metabolic dysfunction. Chemosphere 2019;217:140-9. [Crossref] [PubMed]
- Dumas ME, Kinross J, Nicholson JK. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology 2014;146:46-62. [Crossref] [PubMed]
- Kalhan SC, Guo L, Edmison J, et al. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism 2011;60:404-13. [Crossref] [PubMed]
- Ramirez T, Strigun A, Verlohner A, et al. Prediction of liver toxicity and mode of action using metabolomics in vitro in HepG2 cells. Arch Toxicol 2018;92:893-906. [Crossref] [PubMed]
- Chen Y, Manna SK, Golla S, et al. Glutathione deficiency-elicited reprogramming of hepatic metabolism protects against alcohol-induced steatosis. Free Radic Biol Med 2019;143:127-39. [Crossref] [PubMed]
- Caldez MJ, Van Hul N, Koh HWL, et al. Metabolic Remodeling during Liver Regeneration. Dev Cell 2018;47:425-38.e5. [Crossref] [PubMed]
- Holmes E, Wijeyesekera A, Taylor-Robinson SD, et al. The promise of metabolic phenotyping in gastroenterology and hepatology. Nat Rev Gastroenterol Hepatol 2015;12:458-71. [Crossref] [PubMed]
- Lian JS, Liu W, Hao SR, et al. A serum metabolomic analysis for diagnosis and biomarker discovery of primary biliary cirrhosis and autoimmune hepatitis. Hepatobiliary Pancreat Dis Int 2015;14:413-21. [Crossref] [PubMed]
- Trottier J, Bialek A, Caron P, et al. Metabolomic profiling of 17 bile acids in serum from patients with primary biliary cirrhosis and primary sclerosing cholangitis: a pilot study. Dig Liver Dis 2012;44:303-10. [Crossref] [PubMed]
- Wang JB, Pu SB, Sun Y, et al. Metabolomic Profiling of Autoimmune Hepatitis: The Diagnostic Utility of Nuclear Magnetic Resonance Spectroscopy. J Proteome Res 2014;13:3792-801. [Crossref] [PubMed]
- Yu M, Zhu Y. Metabonomics Research Progress on Liver Diseases. Can J Gastroenterol Hepatol 2017;2017:8467192. [Crossref] [PubMed]
- Rosen CE, Palm NW. Functional Classification of the Gut Microbiota: The Key to Cracking the Microbiota Composition Code: Functional classifications of the gut microbiota reveal previously hidden contributions of indigenous gut bacteria to human health and disease. Bioessays 2017.39. [Crossref] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. [Crossref] [PubMed]
- Yuan J, Chen C, Cui J, et al. Fatty Liver Disease Caused by High-Alcohol-Producing Klebsiella pneumoniae. Cell Metab 2019;30:1172. [Crossref] [PubMed]
- Perry RJ, Peng L, Barry NA, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 2016;534:213-7. [Crossref] [PubMed]
- Hov JR, Karlsen TH. The Microbiome in Primary Sclerosing Cholangitis: Current Evidence and Potential Concepts. Semin Liver Dis 2017;37:314-31. [Crossref] [PubMed]
- Nakamoto N, Sasaki N, Aoki R, et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol 2019;4:492-503. [Crossref] [PubMed]
- O'Hara SP, Karlsen TH, LaRusso NF. Cholangiocytes and the environment in primary sclerosing cholangitis: where is the link? Gut 2017;66:1873-7. [Crossref] [PubMed]
- Liwinski T, Zenouzi R, John C, et al. Alterations of the bile microbiome in primary sclerosing cholangitis. Gut 2020;69:665-72. [Crossref] [PubMed]
- Kunzmann LK, Schoknecht T, Poch T, et al. Monocytes as potential mediators of pathogen-induced Th17 differentiation in patients with primary sclerosing cholangitis (PSC). Hepatology 2020. [Epub ahead of print]. [Crossref] [PubMed]
- Aizarani N, Saviano A. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 2019;572:199-204. [Crossref] [PubMed]
- Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature 2007;449:804-10. [Crossref] [PubMed]
- Zeevi D, Korem T, Zmora N, et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015;163:1079-94. [Crossref] [PubMed]
- Cristoferi L, Nardi A, Ronca V, et al. Prognostic models in primary biliary cholangitis. J Autoimmun 2018;95:171-8. [Crossref] [PubMed]
- Perlman RL. Mouse models of human disease: An evolutionary perspective. Evol Med Public Health 2016;2016:170-6. [Crossref] [PubMed]
- Guo L, Dial S, Shi L, et al. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab Dispos 2011;39:528-38. [Crossref] [PubMed]
- Torres S, Abdullah Z, Brol MJ, et al. Recent Advances in Practical Methods for Liver Cell Biology: A Short Overview. Int J Mol Sci 2020.21. [Crossref] [PubMed]
- Fiorotto R, Amenduni M, Mariotti V, et al. Liver diseases in the dish: iPSC and organoids as a new approach to modeling liver diseases. Biochim Biophys Acta Mol Basis Dis 2019;1865:920-8. [Crossref] [PubMed]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663-76. [Crossref] [PubMed]
- Staerk J, Dawlaty MM, Gao Q, et al. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell 2010;7:20-4. [Crossref] [PubMed]
- Zhou T, Benda C, Duzinger S, et al. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol 2011;22:1221-8. [Crossref] [PubMed]
- Wiegand C, Banerjee I. Recent advances in the applications of iPSC technology. Curr Opin Biotechnol 2019;60:250-8. [Crossref] [PubMed]
- Maepa SW, Ndlovu H. Advances in generating liver cells from pluripotent stem cells as a tool for modeling liver diseases. Stem Cells 2020;38:606-12. [Crossref] [PubMed]
- Si-Tayeb K, Noto FK, Nagaoka M, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 2010;51:297-305. [Crossref] [PubMed]
- Song Z, Cai J, Liu Y, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res 2009;19:1233-42. [Crossref] [PubMed]
- Touboul T, Hannan NR, Corbineau S, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 2010;51:1754-65. [Crossref] [PubMed]
- Corbett JL, Duncan SA. iPSC-Derived Hepatocytes as a Platform for Disease Modeling and Drug Discovery. Front Med (Lausanne) 2019;6:265. [Crossref] [PubMed]
- Rashid ST, Corbineau S, Hannan N, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest 2010;120:3127-36. [Crossref] [PubMed]
- Yusa K, Rashid ST, Strick-Marchand H, et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature 2011;478:391-4. [Crossref] [PubMed]
- Tafaleng EN, Chakraborty S, Han B, et al. Induced pluripotent stem cells model personalized variations in liver disease resulting from α1-antitrypsin deficiency. Hepatology 2015;62:147-57. [Crossref] [PubMed]
- Choi SM, Kim Y, Shim JS, et al. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology 2013;57:2458-68. [Crossref] [PubMed]
- Satoh D, Maeda T, Ito T, et al. Establishment and directed differentiation of induced pluripotent stem cells from glycogen storage disease type Ib patient. Genes Cells 2013;18:1053-69. [Crossref] [PubMed]
- Zhang S, Chen S, Li W, et al. Rescue of ATP7B function in hepatocyte-like cells from Wilson's disease induced pluripotent stem cells using gene therapy or the chaperone drug curcumin. Hum Mol Genet 2011;20:3176-87. [Crossref] [PubMed]
- Jing R, Corbett JL, Cai J, et al. A Screen Using iPSC-Derived Hepatocytes Reveals NAD(+) as a Potential Treatment for mtDNA Depletion Syndrome. Cell Rep 2018;25:1469-84.e5. [Crossref] [PubMed]
- Fiorotto R, Amenduni M, Mariotti V, et al. Src kinase inhibition reduces inflammatory and cytoskeletal changes in DeltaF508 human cholangiocytes and improves cystic fibrosis transmembrane conductance regulator correctors efficacy. Hepatology 2018;67:972-88. [Crossref] [PubMed]
- Ogawa M, Ogawa S, Bear CE, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol 2015;33:853-61. [Crossref] [PubMed]
- Sampaziotis F, de Brito MC, Madrigal P, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol 2015;33:845-52. [Crossref] [PubMed]
- Lorent K, Gong W, Koo KA, et al. Identification of a plant isoflavonoid that causes biliary atresia. Sci Transl Med 2015;7:286ra67. [Crossref] [PubMed]
- Soroka CJ, Assis DN, Alrabadi LS, et al. Bile-Derived Organoids From Patients With Primary Sclerosing Cholangitis Recapitulate Their Inflammatory Immune Profile. Hepatology 2019;70:871-82. [Crossref] [PubMed]
- Ouchi R, Togo S, Kimura M, et al. Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab 2019;30:374-84.e6. [Crossref] [PubMed]
- Cayo MA, Cai J, DeLaForest A, et al. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology 2012;56:2163-71. [Crossref] [PubMed]
- De Assuncao TM, Sun Y, Jalan-Sakrikar N, et al. Development and characterization of human-induced pluripotent stem cell-derived cholangiocytes. Lab Invest 2015;95:1218. [Crossref] [PubMed]
- Dianat N, Dubois-Pot-Schneider H, Steichen C, et al. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology 2014;60:700-14. [Crossref] [PubMed]
- Cayo MA, Mallanna SK, Di Furio F, et al. A Drug Screen using Human iPSC-Derived Hepatocyte-like Cells Reveals Cardiac Glycosides as a Potential Treatment for Hypercholesterolemia. Cell Stem Cell 2017;20:478-89.e5. [Crossref] [PubMed]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347-55. [Crossref] [PubMed]
- Gunther C, Brevini T, Sampaziotis F, et al. What gastroenterologists and hepatologists should know about organoids in 2019. Dig Liver Dis 2019;51:753-60. [Crossref] [PubMed]
- Prior N, Inacio P, Huch M. Liver organoids: from basic research to therapeutic applications. Gut 2019;68:2228-37. [Crossref] [PubMed]
- Zhang RR, Koido M, Tadokoro T, et al. Human iPSC-Derived Posterior Gut Progenitors Are Expandable and Capable of Forming Gut and Liver Organoids. Stem Cell Reports 2018;10:780-93. [Crossref] [PubMed]
- Huch M, Gehart H, van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160:299-312. [Crossref] [PubMed]
- Hu H, Gehart H, Artegiani B, et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018;175:1591-606.e19. [Crossref] [PubMed]
- Nantasanti S, Spee B, Kruitwagen HS, et al. Disease Modeling and Gene Therapy of Copper Storage Disease in Canine Hepatic Organoids. Stem Cell Reports 2015;5:895-907. [Crossref] [PubMed]
- Baktash Y, Madhav A, Coller KE, et al. Single Particle Imaging of Polarized Hepatoma Organoids upon Hepatitis C Virus Infection Reveals an Ordered and Sequential Entry Process. Cell Host Microbe 2018;23:382-94.e5. [Crossref] [PubMed]
- Nie YZ, Zheng YW, Miyakawa K, et al. Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine 2018;35:114-23. [Crossref] [PubMed]
- Schneeberger K, Sanchez-Romero N, Ye S, et al. Large-scale Production of LGR5-positive Bipotential Human Liver Stem Cells. Hepatology 2020;72:257-70. [Crossref] [PubMed]
- Takebe T, Sekine K, Enomura M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013;499:481-4. [Crossref] [PubMed]
- Takebe T, Sekine K, Kimura M, et al. Massive and Reproducible Production of Liver Buds Entirely from Human Pluripotent Stem Cells. Cell Rep 2017;21:2661-70. [Crossref] [PubMed]
- Koike H, Iwasawa K, Ouchi R, et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature 2019;574:112-6. [Crossref] [PubMed]
- Liu HY, Lin CC. A Diffusion-Reaction Model for Predicting Enzyme-Mediated Dynamic Hydrogel Stiffening. Gels 2019;5:17. [Crossref] [PubMed]
Cite this article as: Oliveira AG, Fiorotto R. Novel approaches to liver disease diagnosis and modeling. Transl Gastroenterol Hepatol 2021;6:19.


