
Proteomic heterogeneity reveals SOAT1 as a potential biomarker for hepatocellular carcinoma
Liver cancer is one of the few and fastest neoplastic malignancy to continue to rise in incidence in the United States and across the world (1). Today, the 5-year survival rate of liver cancer is approximately 17%, making it the 2nd most common cause of death from cancer worldwide (2). A highly heterogeneous disease, hepatocellular carcinoma (HCC) is the more common subtype of liver cancer and research lacks information on the prominent drivers that promote disease progression leading to limited therapeutic options. However, common yet diverse risk factors have been identified that promote tumors heterogeneity which include Hepatitis B or C viral infection, liver flukes, diet, and alcohol. Together, these factors lead to cirrhosis of the liver, ultimately putting individuals at risk of developing liver cancer. Because there is a sequential nature of the onset of HCC with liver cirrhosis as the main risk factor, scientists and clinicians have sought to determine if HCC can be detected at an early stage of carcinogenesis before advancing to the later, more deadly stage of disease. Advances in omic-based technologies have made tremendous efforts in unmasking genotypic and phenotypic features of HCC such as transcriptomics, genomics, metabolomics, and proteomics. While these approaches give in depth analyses of tumors with multiple and diverse cellular pathways coming forward as contributors of cancer development, it is difficult to parse through which gene hits are the key players of hepatocarcinogenesis to further validate in functional studies.
For the past 10 years, whole exome sequencing, RNA-sequencing, and array-based platforms have been the gold standard in deciphering the genomic architecture of tumors with single-cell RNA sequencing emerging as the most in-depth analysis to study cancer at the single cell level. However, these analyses only stop at the mRNA processing of the central dogma of biology and does not give a complete picture of the structural and functional processes of proteins that ultimately lead to phenotypic observations. Thus, the field of proteomics has emerged to unveil the knowledge of the processes involved in DNA to protein to phenotype interactions to gain greater insight into disease biology. Mass spectrometry (MS) is the main technology implored to identify proteins, peptides, and posttranslational modifications with quantitative and qualitative measures of protein levels, protein structures, protein-protein interactions, and protein-nucleic acid interactions. Liquid chromatography has been coupled with mass spectrometry (LC-MS or LC-MS/MS) to increase sensitivity while minimizing background noise. To date, approximately eight studies have been published combining genomic and proteomic analyses in human tumor samples, summarized in Table 1, to better identify biomarkers of tumor development for high-risk patients that is not evident at the mRNA level (3-10). Thus, proteomics is a useful technology to gain a greater depth of knowledge of cancer biology and may be essential in drug development for translational studies.
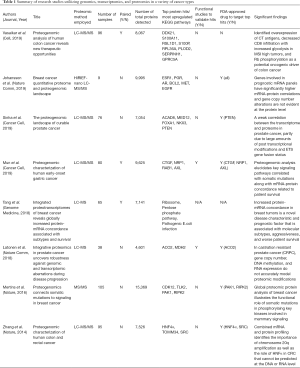
Full table
Recently, a paper published in Nature titled, “Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma,” Jiang et al., sought to investigate the proteomic landscape of patients (110 paired tumor and nontumor clinical samples) infected with Hepatitis B virus and whether there are certain proteins linking to early-stage HCC. Their comprehensive proteomic data elegantly revealed extensive heterogeneity amongst their samples, allowing them to stratify patients using a non-negative matrix factorization consensus-clustering into three subtypes, S-I, S-II, S-III, each illustrating different clinical outcomes as well as unique signature proteins (11). Patients consisting in the S-III subtype displayed the worse overall survival as well as higher HCC recurrence, which may in part be contributed to the higher levels of microscopic vascular invasion (MVI) and alpha-fetoprotein (AFP) compared to the S-I and S-II subtypes. Because the S-III subtype has the poorest prognosis, the authors developed prognostic risk scores for the most abundant and FDA-approved or under clinical trials drug-targetable proteins in the S-III patients. They identified sterol O-acyltransferase 1 (SOAT1) with the highest risk score for a mortality prognosis of HCC. SOAT1 is involved in the formation of cholesterol esters from cholesterol and its expression was also elevated in a tissue microarray analysis as well as correlated with overall survival in The Cancer Genome Altas (TCGA), independent of other risk factors. Moving forward, in order to validate SOAT1 as a potential therapeutic target, the authors conducted various functional assays including short hairpin knockdown of SOAT1 in two HCC cell lines, treatment of cells with avasimibe, a SOAT1 inhibitor, and avasimibe-targeted SOAT1 inhibition in patient-derived tumor xenograft (PDX) models of HCC. Collectively, a reduction or inhibition of SOAT1 significantly reduced cellular proliferation, migration, tumor growth, and potential for metastasis. We believe these findings are instrumental additions to the field of liver cancer research and exemplify much needed knowledge for the role of proteins in the onset of HCC.
Due to the rising rates of liver cancer, identifying new and novel therapeutic targets of hepatocellular carcinoma is imperative. One way to further our understanding of the mechanisms that promote carcinogenesis is through analysis of the proteome. A key theme that emerges in proteogenomic/proteotranscriptomic studies is that proteomics identifies differentially regulated proteins that cannot be depicted at the DNA or RNA level. Thus, we believe the novelty of this paper is that it utilizes proteomic analysis, whole-genome sequencing, and RNA sequencing to identify clinically targetable proteins that are elevated in paired samples of early-stage HCC. This is the first study in HCC to date, that performs a system-wide approach to unmask DNA to protein to phenotype changes in liver cancer tissue with the identification of SOAT1 as a potential biomarker for early-stage HCC. Having a role in the production of converting endoplasmic reticulum cholesterol to cholesterol esters to then be stored in lipid droplets, SOAT1 has only been studied exclusively, in the context of cancer, in glioblastoma (GBM), prostate, and breast cancer. Geng et al. showed that inhibition of SOAT1 reduces GBM growth and increases survival via suppression of sterol regulatory element-binding protein-1 (SREBP-1)-mediated lipid synthesis in a xenograft model (12). Therefore, it is sufficient to say that SOAT1 is a new therapeutic target in not just HCC but potentially in other cancer types. Their results leading to SOAT1 was further encouraging when an already FDA-approved drug that can inhibit its function in cholesterol ester synthesis, avasimibe, is available for current medicinal practices today, adding an additional novelty to the paper.
Alongside their finding of SOAT1, the authors further validated their bioinformatic results with functional assays in in vitro and in vivo models, an addition that has been lacking in other proteogenomic studies. Conducting multiple functional assays, their in vitro and in vivo results were able to recapitulate their proteomic findings, strengthening their claims that SOAT1 can be a potential therapeutic biomarker for HCC. Moreover, another methodical asset this paper provides is the use of liquid chromatography with tandem mass spectrometry (LC-MS/MS), which is highly more specific and sensitive to the other commonly used method, LC-MS, making the results more robust and reproducible. By having an additional fragmentation step, proteins of similar masses are able to be separate into different entities, leading to lower false discovery rates than LC-MS. Thus, we believe the findings presented in the paper can be accepted with a greater confidence than other proteogenomic studies that fail to show a functional validation of their top proteomic hits.
It is worth noting that the authors describe little to no information on whether there exists mRNA-protein concordance possibly due to the lack of transcriptome data for most of the tumor samples (there is a limited number of overlapping samples that have both transcriptomic and proteomic data). Numerous studies infer changes in functional protein by mRNA abundance, which is largely inaccurate and misleading. The studies reported in Table 1 integrate genomic and proteomic data to a greater extent than this manuscript and describe new proteomic inferences that are not detected at the genomic level or misinformation at the DNA or RNA level. For example, Vasaikar et al. combines genomic and proteomic data of colon cancer to demonstrate that based on somatic mutation data, SOX9 was predicted to be a tumor suppressor, however proteomic analysis revealed it to be an oncogene. Similarly, their phosphoproteomic data identified Rb phosphorylation as a potential oncogenic driver of colon cancer, which previously was not discovered using only genomic and transcriptomic data (3). Moreover, Sinha et al. further illustrates these points as increased protein abundance of PUS1 in prostate cancer patient is associated with an increase risk of biochemical relapse (BCR) whereas increased mRNA abundance correlates with a reduced risk (5). The notion that genomic and transcriptomic profiles of tumor samples are a surrogate for their proteomic landscape may be insufficient to claim based on the studies depicted above. To further expand on this point, the authors do not do subtype analysis on their transcriptomic data (as well as relation to other subtypes of HCC from TCGA data) and only focus on their proteomic data. Thus, integrating similar analyses from transcriptomic, genomic, proteomic, and metabolic data from the same tumor samples through potentially iCluster (an integrative clustering framework) can provide new insights and a more comprehensive depiction of cancer biology towards greater biomarker discoveries (13).
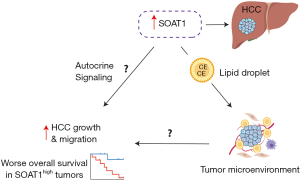
Moreover, the authors show that in their proteomics data, there is a greater number of SOAT1 low tumors vs. high and only high SOAT1 PDX’s have a response to avasimibe with a significant reduction in tumor growth at day 28. However, it would be worth having a graph depicting the mRNA expression of SOAT1 in HCC tumors vs. nontumor from their samples to determine whether this is a universal biomarker to target for most if not all liver cancer patients. This would also further elaborate whether there is a concordance of mRNA and protein expression of SOAT1 as the S-III proteomic subtype expresses the highest amount of this biomarker. Likewise, it is difficult to assess the universality of SOAT1 for the future without having conducted a clinical trial investigating treatment of avasimibe in HCC patients. It is not known whether a clinical trial has been initiated to treat early-stage HCC patients with avasimibe, which if proven to be effective can make a monumental contribution to the current standard of care given to these patients. Furthermore, it is not made clear how regulating the levels of cholesterol esters via avasimibe reduces tumor growth and only briefly mentions that cholesterol homeostasis is modified in HCC with no further explanation and functional studies (proposed mechanism in Figure 1).
With the wave of immunotherapy and the lack of knowledge on why HCC patients fail to respond to treatment that implores the use of the immune system, identifying the drivers that promote reduced immunosurveillance and resistance to immune checkpoint inhibitors is warranted. In the paper being described, the authors mention little information on the proteome and the immune system. They make brief statements on increased immune infiltration in the S-III subtype, corresponding to enrichment of T-exhaustion, immunosuppressive regulatory T-cell, and M2-macrophage signatures. It would be worth providing more information on the interaction of proteins and immune cells to determine whether there are different observations portrayed compared to our current knowledge of the immune system through DNA and RNA analyses. Furthermore, it would also be interesting to know whether these tumors are high or low in microsatellite instability. Recent evidence has shown that microsatellite instability high tumors are better equipped to respond to immunotherapy than their counterpart, illustrating a potential mechanism to further investigate (14).
Currently, single cell RNA-sequencing has become one of the most exciting technologies developed and implemented in the past 5 years. By being able to identify the genomic makeup of cells at the single cell level, researchers and clinicians have gain a greater understanding of cancer biology, inter and intratumoral heterogeneity, and therapy resistance. In particular, single-cell analysis has revealed cancer stem cell heterogeneity in HCC (15). However, current single cell technologies have yet to create a platform to investigate proteins at the single level, causing proteomic analyses today to continue to fall behind that of DNA and RNA. If we are wanting to make significant discoveries in biomedical research, we must establish streamlined pipelines that integrate all steps of the central dogma of biology and not infer that the observations seen in DNA and RNA can be translated to protein function. The field of proteomics is just beginning to increase in interest in cancer research and must be further implemented in all cancer types to prove its usefulness which will then encourage scientists to develop assays to detect and measure single proteins. In turn, this will lead to greater cancer biomarker discoveries as it is an exciting and clinically translatable field that can provide greater therapeutic options to improve patients’ lives. Jiang et al. demonstrate the utility of proteomics in detecting various altered proteins and are the first pioneers to conduct this analysis in HCC, opening the door to future therapeutic opportunities.
Acknowledgments
This work was supported by the intramural program of the Center for Cancer Research, National Cancer Institute.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Cronin KA, Lake AJ, Scott S, et al. Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics. Cancer 2018;124:2785-800. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Vasaikar S, Huang C, Wang X, et al. Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities. Cell 2019;177:1035-49.e19. [Crossref] [PubMed]
- Johansson HJ, Socciarelli F, Vacanti NM, et al. Breast cancer quantitative proteome and proteogenomic landscape. Nat Commun 2019;10:1600. [Crossref] [PubMed]
- Sinha A, Huang V, Livingstone J, et al. The Proteogenomic Landscape of Curable Prostate Cancer. Cancer Cell 2019;35:414-27.e6. [Crossref] [PubMed]
- Mun DG, Bhin J, Kim S, et al. Proteogenomic Characterization of Human Early-Onset Gastric Cancer. Cancer Cell 2019;35:111-24.e10. [Crossref] [PubMed]
- Tang W, Zhou M, Dorsey TH, et al. Integrated proteotranscriptomics of breast cancer reveals globally increased protein-mRNA concordance associated with subtypes and survival. Genome Med 2018;10:94. [Crossref] [PubMed]
- Latonen L, Afyounian E, Jylha A, et al. Integrative proteomics in prostate cancer uncovers robustness against genomic and transcriptomic aberrations during disease progression. Nat Commun 2018;9:1176. [Crossref] [PubMed]
- Mertins P, Mani DR, Ruggles KV, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016;534:55-62. [Crossref] [PubMed]
- Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014;513:382-7. [Crossref] [PubMed]
- Jiang Y, Sun A, Zhao Y, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 2019;567:257-61. [Crossref] [PubMed]
- Geng F, Cheng X, Wu X, et al. Inhibition of SOAT1 Suppresses Glioblastoma Growth via Blocking SREBP-1-Mediated Lipogenesis. Clin Cancer Res 2016;22:5337-48. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41.e23. [Crossref] [PubMed]
- Mandal R, Samstein RM, Lee K-W, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti–PD-1 immunotherapy response. Science 2019;364:485-91. [Crossref] [PubMed]
- Zheng H, Pomyen Y, Hernandez MO, et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 2018;68:127-40. [Crossref] [PubMed]
Cite this article as: Khatib SA, Wang XW. Proteomic heterogeneity reveals SOAT1 as a potential biomarker for hepatocellular carcinoma. Transl Gastroenterol Hepatol 2019;4:37.


